 Open Access
Open Access
ARTICLE
Anti-cancer effects of sitagliptin, vildagliptin, and exendin-4 on triple-negative breast cancer cells via mitochondrial modulation
1 School of Biotechnology, Devi Ahilya University, Indore, 452001, India
2 Laboratory of Membrane Protein Biology, National Centre for Cell Science, Pune, 411007, India
3 Department of Biosciences and Biomedical Engineering, Indian Institute of Technology, Indore, 452020, India
4 Department of Biotechnology, IPS Academy, Indore, 452012, India
5 School of Pharmacy, Devi Ahilya University, Indore, 452001, India
6 Central Council for Research in Ayurvedic Sciences, Kolkata, 263645, India
* Corresponding Author: HAMENDRA SINGH PARMAR. Email:
BIOCELL 2022, 46(12), 2645-2657. https://doi.org/10.32604/biocell.2022.021754
Received 02 February 2022; Accepted 13 April 2022; Issue published 10 August 2022
Abstract
Triple-negative breast cancer (TNBC) cell line MDA-MB-231 is known for Warburg metabolism and defects in mitochondria. On the other hand, dipeptidyl peptidase-IV (DPP-IV) inhibitors such as sitagliptin and vildagliptin and GLP-1 agonist exendin-4 are known to improve mitochondrial functions as well as biogenesis, but no study has evaluated the influence of these drugs on mitochondrial biogenesis on metastatic breast cancer cell line. We have recently reported anticancer effects of 5-aminoimidazole-4-carboxamide riboside on MDA-MB-231 cells via activation of AMP-dependent kinase (AMPK), which activates the downstream transcription factors PGC-1α, PGC-1β, or FOXO1 for mitochondrial biogenesis; above-mentioned incretin-based therapies are also known to activate AMPK. This study evaluated the effects of sitagliptin, vildagliptin, and exendin-4 on MDA-MB-231 cells and the underlying changes in mitochondrial biogenesis, were examined. Treatment with sitagliptin (100 µM), vildagliptin (100 µM), and exendin-4 (10 nM) for 72 h to MDA-MB-231 cells led to a decrease in viability indicated by MTT assay, cell migration by scratch, and transwell migration assays, accompanied with marginal reduction in cell numbers along with the apoptotic appearance, the rate of apoptosis, and decreased lactate content in conditioned medium. These changes in the cancer phenotype were accompanied by an increase in the mitochondrial DNA to nuclear DNA ratio, increased MitoTracker green and red staining, and increased expression of transcription factors PGC-1α, NRF-1, NRF-2, TFAM, and HO-1. Pre-treatment of cells with these incretin-based drugs followed by 48 h treatment with 1 µM doxorubicin increased doxorubicin sensitivity as observed by a decrease in viability by MTT assay. Thus, sitagliptin, vildagliptin, and exendin-4 exert their beneficial effects on TNBC cells via an increase in mitochondrial biogenesis that helps to switch Warburg metabolism into anti-Warburg effect. Therapeutic response was in the order of: sitagliptin > vildagliptin > exendin-4.Keywords
Mitochondrial defects have been commonly reported in most cancer cells, and thus, metabolic alterations are thought to play an important role in carcinogenesis (Rouzier et al., 2005; Carey et al., 2006). Aerobic glycolysis or Warburg metabolism is one of the hallmarks of almost all the cancers contributing to the proliferation and inhibition of cancer cell death (Fadaka et al., 2017; Avagliano et al., 2019). Depletion in mitochondrial number and mutations in mitochondrial DNA has been reported in breast cancer (Tseng et al., 2006). Additionally, a decrease in mitochondrial number has been correlated with the tumor progression and poor prognosis in patients with breast cancer (Yu et al., 2007).
Breast cancer cells, including the most aggressive triple-negative breast cancer cells (TNBC) as well as cancer-initiating cells, are also characterized by Warburg-type metabolic rearrangements, reduced mitochondrial activity, and uncoupling (Ayyasamy et al., 2011; Kim et al., 2013; Feng et al., 2014; Tang et al., 2014). Interestingly, anti-Warburg rearrangements are known to support chemotherapeutic responses in breast cancer (Suhane and Ramanujan, 2011).
Recently, we have comprehensively reviewed the crosstalk between diabetes and breast cancer pathways and highlighted the importance of the repurposing of incretin-based antidiabetic therapies in breast cancer (Parmar et al., 2021; Parmar et al., 2022; Jaiswal et al., 2022).
Incretins are a group of metabolic hormones known to potentiate insulin secretion in response to glucose or food stimuli. Glucagon-like peptide-1 (GLP-1) is an incretin, which is formed by the posttranslational processing of proglucagon gene transcript in intestinal L-cells of the gastrointestinal tract (GIT) and exerts glucose-lowering effects via binding with GLP-1 receptors. Incretins exert pleiotropic antidiabetic effects, including insulin secretion in response to glucose, diminished glucagon secretion, delayed gastric emptying, reduced hepatic glucose output, and increased pancreatic β-cell survival (Jaiswal et al., 2022). However, the metabolic fate of GLP-1 is determined by an enzyme known as dipeptidyl peptidase-IV (DPP-IV), which is a subset of oligopeptidase encoded by CD26. It is widely distributed in various tissues. DPP-IV enzyme cleaves off dipeptides from the peptides having alanine or proline at position 2 of their N-terminal. Therefore, it inactivates the incretin and promotes diabetes. Interestingly, incretin-based antidiabetic therapies, including GLP-1 agonists and DPP-IV inhibitors, are known to exert pancreatic as well as extra pancreatic effects; some of these pathways have indicated their potential in the treatment of breast cancer (Jaiswal et al., 2022).
On the one hand, TNBC and cancer-initiating cells are characterized by Warburg-type metabolic rearrangements (Ayyasamy et al., 2011; Kim et al., 2013; Feng et al., 2014; Tang et al., 2014); on the other hand, incretin-based therapies have been shown to promote mitochondrial biogenesis and improvement in mitochondrial metabolism, suggesting the potential of these therapies in breast cancer treatment. The DPP-IV inhibitor drug sitagliptin promotes mitochondrial biogenesis via increasing the expression of PGC-1α, NRF-1, and TFAM and improving mitochondrial functioning (Apaijai et al., 2013; Abuelezz et al., 2016; Weng et al., 2019). Similarly, vildagliptin is reported to promote the expression of the genes regulating mitochondrial biogenesis (Yang et al., 2020). A GLP-1 agonist, exendin-4, also promotes mitochondrial biogenesis via PGC1-α, sirtuin 1 (SIRT-1), activation of AMP-activated protein kinase (AMPK), and c-Jun-N-terminal kinase (JNK) proteins (Kang et al., 2015; Strycharz et al., 2018; Mostafa Tork et al., 2019; Igoillo-Esteve et al., 2020). Importantly, AMPK activation is known to upregulate the mitochondrial biogenesis pathway (Tripathi et al., 2022).
In our recent review on the therapeutic potential of repurposing the incretin-based therapies in breast cancer, we explored all the plausible molecular mechanism(s) of DPP-IV inhibitors and GLP-1 agonists along with the cancer-promoting role of DPP-IV (Jaiswal et al., 2022). Recently, we also reported the anti-cancer role of AMPK activator, 5-aminoimidazole-4-carboxamide-1-β-D-ribofuranoside (AICAR) via increased expression of mitochondrial biogenesis related genes PGC-1α, NRF-1, NRF-2, and TFAM (Tripathi et al., 2022) which is in line with the reports available on the AMPK activation by sitagliptin, vildagliptin and exendin-4 (Zheng et al., 2018; Nath et al., 2021; Li et al., 2021). These reports provided another clue for the therapeutic potential of sitagliptin, vildagliptin, and exendin-4 on breast cancer, possibly via the promotion of mitochondrial biosynthesis. However, no report is available on these incretin-based therapies on breast cancer cells exploring the changes in mitochondrial biogenesis and possible reversal of aerobic glycolysis or Warburg-type metabolism associated with the therapeutic response on breast cancer. Therefore, in the present study, we examined the effects of sitagliptin, vildagliptin, and exendin-4 on TNBC cells MDA-MB-231 by evaluating viability, apoptosis, migration, proliferation as well as changes in mitochondrial biosynthesis-related parameters including mitochondrial (mt)-DNA to nuclear (n)DNA ratio, MitoTracker green and red staining for mitochondrial biomass and membrane potential, respectively, and lactate estimation in conditioned media as a marker of changes in the glycolytic pathway. Importantly, we also studied the influence of pretreatment with the test drugs on the sensitivity of doxorubicin by viability assay.
Dulbecco’s modified eagle medium (DMEM), fetal bovine serum (FBS), Dulbecco’s phosphate-buffered saline (DPBS), TrypLETMExpress enzyme, and antibiotics (Penicillin-streptomycin) were purchased from Gibco Thermo Fisher Scientific. Doxorubicin, sitagliptin phosphate vildagliptin, exendin-4, thiazolyl blue tetrazolium bromide (MTT), and cell death detection kits were purchased from Sigma-Aldrich, Pvt., Ltd., Mo, USA. The lactate colorimetric assay kit was purchased from Bio Vision, Milpitas, USA. Sitagliptin and exendin-4 were dissolved in molecular grade distilled water, whereas vildagliptin was initially dissolved in a trace amount of dimethyl sulfoxide (DMSO), and then dilutions were prepared with molecular grade distilled water. Aliquots of these drugs were stored at −20°C. For experiments, stock solutions were further diluted in a serum-free culture medium.
Chemical formulae and structures of sitagliptin, vildagliptin, and exendin-4
Sitagliptin phosphate monohydrate (C16H15F6N5O · H3PO4 · H2O)
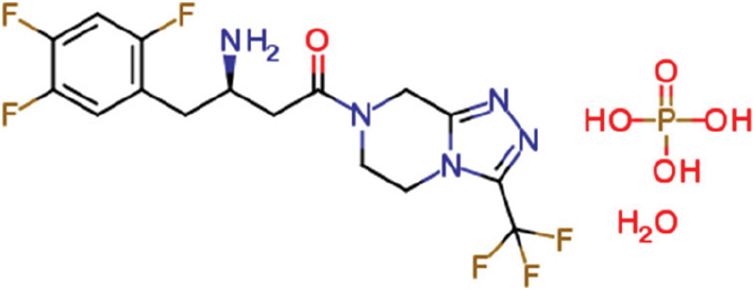
Mechanism of action: Sitagliptin selectively binds with the DPP-IV enzyme, through which it inhibits DPP-IV activity that prevents degradation of GLP-1 and other GIPs that exert pleiotropic antidiabetic effects such as an increase in insulin secretion and pancreatic β-cell survival, decreased glucagon secretion, delayed gastric emptying and decrease in hepatic glucose output. DPP-IV is known to promote the process of carcinogenesis via collagen breakdown, inhibition of CXCL12 and CXCR4 that diminish anti-tumor functions of T & NK cells, promotion of EGF signaling responsible for cancer cell survival and other oncogenic pathways, enhanced PIN1 expression which promotes epithelial to mesenchymal transition) which increases tumor stemness (Jaiswal et al., 2022). In response to sitagliptin treatment, we observed the promotion of mitochondrial biogenesis and reversal of Warburg-type metabolism, which seems to be an underlying mechanism for its anti-cancer effects on the TNBC cell line.
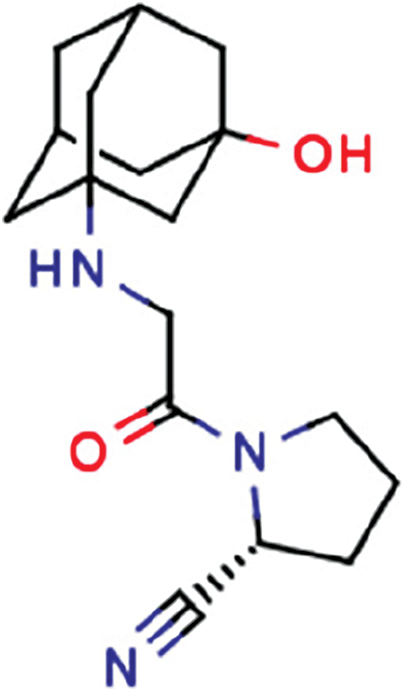
Mechanism of action: Vildagliptin is another selective inhibitor of the DPP-IV enzyme, which prevents degradation of GLP-1 and other GIPs that exert pleiotropic antidiabetic effects as mentioned above for sitagliptin. There are no reports on the anti-cancer effects of vildagliptin, but as DPP-IV activity itself is known to promote carcinogenesis and metastasis in a variety of cancers, including breast cancer, vildagliptin may exert anti-cancer effects through inhibition of DPP-IV (Parmar et al., 2021; Jaiswal et al., 2022; Parmar et al., 2022). In present study, we observed anti-cancer effects of vildagliptin on the TNBC cell line, possibly via mitochondrial biogenesis and reversal of Warburg-like metabolism.
Peptide sequence:
H-His-Gly-Glu-Gly-Thr-Phe-Thr-Ser-Asp-Leu-Ser-Lys-Gln-Met-Glu-Glu-Glu-Ala-Val-Arg-Leu-Phe-Ile-Glu-Trp-Leu-Lys-Asn-Gly-Gly-Pro-Ser-Ser-Gly-Ala-Pro-Pro-Pro-Ser-NH2
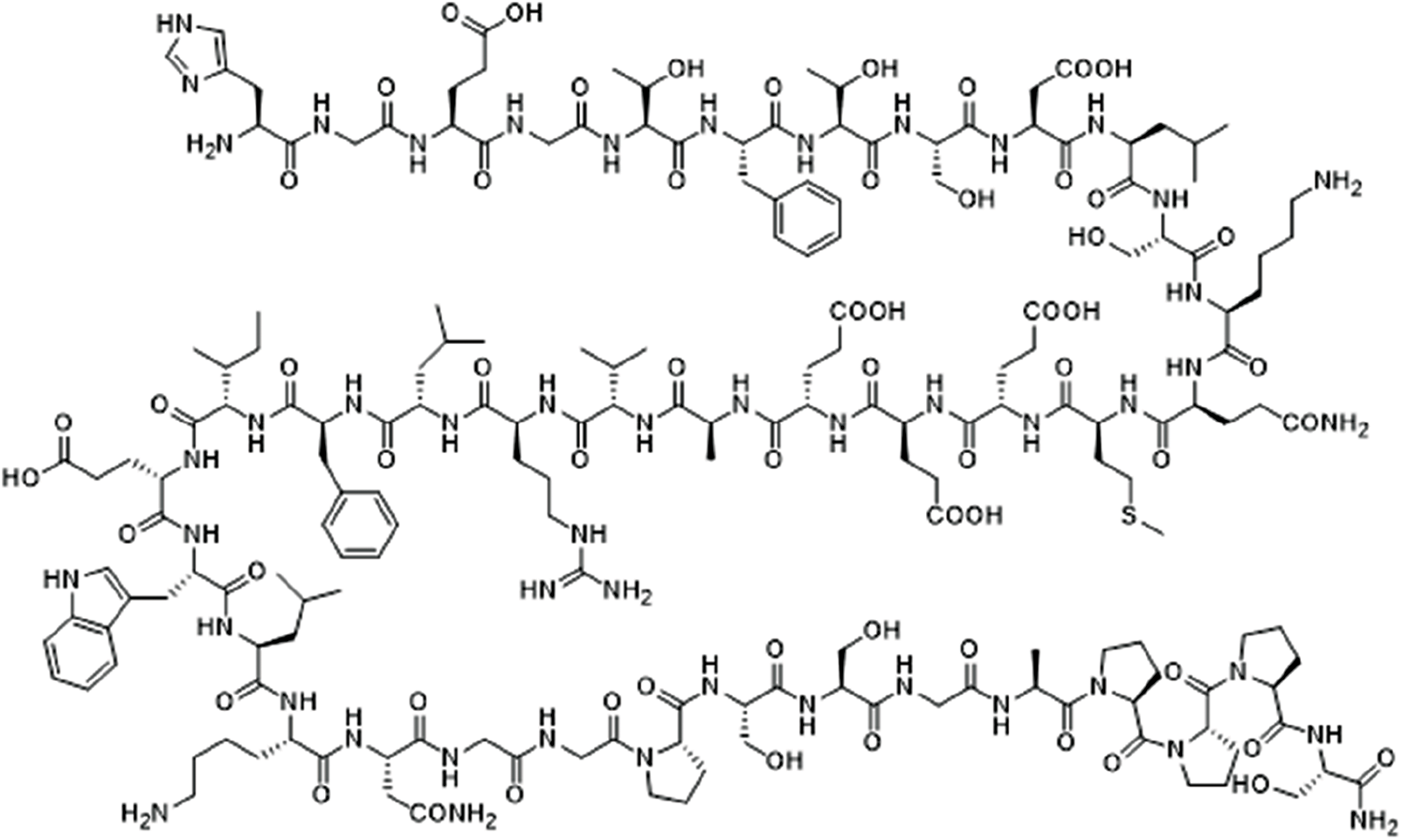
Mechanism of action: Exendin-4 is a GLP-1R agonist that binds with the GLP-1 receptor, which exerts similar antidiabetic effects as observed in the case of DPP-IV inhibitors sitagliptin and vildagliptin (as mentioned above). Besides, exendin-4 exerts its anti-cancer effects via inhibition of PI3K/Akt, MAPK/ ERK, and NF-κβ signaling pathways, increase in insulin sensitivity and anti-inflammatory effects, increase in anti-tumor immune response, stalling pro-survival and proliferative pathways and inducing pro-apoptotic pathways, E-cadherin up-regulation, increased chemosensitivity via p-mTOR, cyclin B and p34cdc2 (Jaiswal et al., 2022). In the present study, we observed anti-cancer effects of exendin-4 on the TNBC cell line, possibly via mitochondrial biogenesis and reversal of Warburg-like metabolism.
Triple-negative breast cancer cell line (MDA-MB-231) was obtained from the cell culture repository of the National Centre for Cell Science (NCCS), Pune, India. Cells were cultured in DMEM supplemented with 10% FBS (v/v) and 1% antibiotics (100 U/mL penicillin and 100 U/mL streptomycin) in a humidified atmosphere of 5% CO2 at 37°C and the media was changed each time after two days interval.
Cell viability was measured by MTT assay. In this assay, 1 × 104 cells/well were seeded into 96-well plates and allowed to adhere properly for 12 h. After adhesion, the cells were treated with different doses of sitagliptin, vildagliptin, and exendin-4, while in control wells; an equal volume of fresh media was added. Media control wells were devoid of the cells containing media without serum. Plates were incubated for 72 h, the media was removed, and wells were washed with DPBS. After adding 180 µL of fresh media, 0.5 µg of the final concentration of MTT reagent (20 µL from 5 mg/mL stock) was added to each well. Plates were kept in a CO2 incubator for 4 h. After incubation, the media was removed carefully, and formazan crystals were dissolved by the addition of a 200 µL mixture of DMSO and ethanol (1:1) into each well. Absorbance was taken at 570 nm with a reference wavelength of 620 nm in enzyme-linked immunosorbent assay (ELISA) plate reader (Microscan- MS5608A, ECIL, Hyderabad, India).
Viability/apoptotic morphology
MDA-MB-231 cells were cultured in a 12-well plate (1 × 105 cells/well) with 1 mL complete DMEM and were allowed to reach 70%–80% confluence. Cells were then treated with all the doses of sitagliptin, vildagliptin, and exendin-4 for 72 h and then observed under the inverted light microscope. The apoptotic phenotype was characterized by morphological distortion and loss of cellular contact with the matrix. An apparent decrease in cell numbers also indicated cell death (Hu et al., 2016; Nasser et al., 2017; Tripathi et al., 2022).
Scratch/wound healing assay
The migratory potential of the cells was determined by a wound-healing assay. Cells were seeded into 24 well plates (5 × 104 cells/well) and allowed to acclimatize overnight. Cells were treated with the selected doses of sitagliptin, vildagliptin, and exendin-4 and incubated for 72 h. Then, the wound was created in each well using a 200 μL pipette tip, and the media was removed. The plate was washed with DPBS to remove cell debris, and these wells were replenished with fresh media. Cell migration was observed at t = 0 and t = 24 h. Distance of the scratch was measured by calculating the width of the wound healed by the migratory cells. The experiment was conducted with n = 5 which is number of wells used for each group and repeated thrice.
Cancer cells exhibit chemotactic movement towards cytokines, proteins, and other factors present in the FBS. For migration assay, 24 well-hanging inserts with 8 μm pore size polyester membranes were used to study chemotactic movements (Falcon, Cat # 353097). Initially, cells were grown in a normal culture plate with complete DMEM media along with the treatment of the selected doses of sitagliptin, vildagliptin, and exendin-4 for 72 h. After trypsinization, 5 × 104 cells/well were counted and reseeded into the insert wells (upper chamber) of the transwell migration plate containing 200 μL of serum-free media, whereas the lower chamber had 600 µL of complete DMEM (10% FBS as a chemo-attractant). Plates were kept in humidified condition (5% CO2) at 37°C for 24 h. After 24 h, non-invading cells were carefully removed using a cotton swab. Cells that migrated towards the lower side of the insert by passing through the insert filter were fixed with 3.7% formaldehyde and permeabilized with 100% methanol. Migratory cells were stained with 0.1% crystal violet, and images were acquired by inverted light microscopy (Leica DM, IL, LED).
Staining using MitoTracker Red and Green was performed following the instructions of the manufacturer. MitoTracker red was used to assess the membrane potential, and MitoTracker Green was used to assess the mitochondrial biomass. Cells were grown on 12 mm round-shaped coverslips placed in 6-well plates. Then, the selected doses of sitagliptin, vildagliptin, and exendin-4 were added in the experimental wells, while in control wells an equal volume of fresh media was added. Then, the plates were incubated for 72 h in a CO2 incubator under appropriate culture conditions. After completion of incubation time, media was removed from the wells, and the cells were washed by DPBS. Fresh serum-free media was added to these wells, and cells were incubated sequentially with MitoTracker Red (300 nM) followed by MitoTracker Green (100 nM) for 25 min at 37°C in a CO2 incubator. The unbound stain was removed by washing twice with a serum-free medium. Cells were fixed by 3.7% formaldehyde and permeabilized by 100% methanol. After washing, 1.5 µL of DAPI (5 µg/ml) was used for nuclear staining for 5 min, followed by washing with DPBS. Antifade mounting media (Vectashield Cat # H-1000-10) was used, and images were captured by confocal microscopy (Zeiss LSM 880 with Airyscan). Fluorescence intensities were quantified by using Image J software.
Cell death detection was determined using an ELISA kit from Merck, Roche Life Science (USA). Briefly, 1 × 104 cells were seeded into 96-well plates initially, and after adherence, cells were incubated with the test drugs, while in control wells, an equal volume of fresh media was added. Media was removed after 72 h of incubation and cells were washed thrice with DPBS. Then, 200 µL of lysis buffer was added to each well and incubated at room temperature (RT) for 30 min and then lysates were collected into microcentrifuge tubes. Tubes were centrifuged at 200 g for 10 min, and 20 µL supernatant from each sample was added to the respective wells of the ELISA plate. Then, 80 µL of immunoreagent was added, and the plate was incubated for 2 h with shaking at room temperature. After incubation, the solution from the wells was discarded, and the wells were washed thrice with incubation buffer. Then, 100 µL of ABTS solution was added, and the plate was placed on a shaker for 30 min at room temperature. Finally, 100 µL of stop solution was added to each well, and the absorbance was measured spectrophotometrically using an ELISA plate reader at 405 nm and a reference wavelength of 490 nm. Mixtures of 100 µL ABTS solution and 100 µL of stop solution were used as a blank.
Cells were treated with the test drugs. Treated and untreated (control) groups were incubated for 72 h in 24 well plates. After 72 h, media was removed, and cells were washed thrice with DPBS. An equal number of cells (1 × 104 cells) were trypsinized and reseeded into a new 96-well plate. DMEM without phenol red and serum was added, and plates were incubated for 24 h. Lactate assay was performed as per the protocol provided by the manufacturer (Biovision, Cat # K951-384). After 24 h, 12.5 µL of media was collected from each well for the detection of lactate. The reaction mixture was prepared using 10 µL of working assay buffer, 0.5 µL of enzyme mix, and 2 µL of the probe per reaction. In this reaction mix, 12.5 µL of conditioned media was added and incubated for 30 min at room temperature, and the absorbance was measured at 590 nm against the blank sample containing 12.5 µL of media without cells.
Relative quantification of mitochondrial DNA using quantitative real-time polymerase chain reaction (qPCR)
To evaluate the relative change in the quantity of mitochondrial DNA, cells were initially grown in the presence or absence of the test drugs for 72 h. Then fresh media was added and cells were further incubated for 48 h in the CO2 incubator. Subsequently, the media was removed and cells were washed thrice using DPBS, followed by trypsinization. Then, 1.0 mL of DPBS was added and cells were pelleted by centrifugation at 1200 rpm for 5 min at room temperature. One more cycle of washing was repeated and DPBS was removed from the pellet. These cells were used for the isolation of total DNA or RNA. DNA was isolated using a pure link genomic DNA mini kit (Invitrogen, Cat # K-1820-01). For qPCR, the Aria Mxreal-time PCR system from Agilent was used. The reaction mixture consisted of 0.4 µL (0.4 µM) forward and 0.4 µL (0.4 µM) reverse primer, 10 µL of 2X SYBR green master mix (Applied biosystems Cat # A25741), 7.8 µL of DW, and 1 µL of DNA sample (4 ng) as PCR template. The sequences of the primers to amplify a region corresponding to the D-loop of human mitochondria were as follows: 5’-GCA GAT TTG GGT ACC ACC CAA GTA TTG ACT CAC CC-3’ (F), 5’-GCA TGG AGA GCT CCC GTG AGT GGT TAA TAG GGT GATAG-3’ (R) and for nuclear DNA, primers for the β-globin gene: 5’-GAAGAGCCAAGGACAGGTAC-3’ (F) and 5’-CAACTTCATCCACGTTCACC-3’(R) were used. For the PCR, the following program was used: 1 cycle at 95°C for 15 min, followed by 40 cycles at 95°C for 30 s, 58°C for 30 s, and 72°C for 90 s. The relative amount of mtDNA to nDNA compared to the control groups was then analyzed using the 2−ΔΔCt formula (Tripathi et al., 2022). Each sample was run in duplicate, and the average Ct value was considered for the calculation of relative fold change.
Quantitative gene expression analysis
The cells were treated with the test drugs and harvested in the same manner as mentioned above for DNA isolation. RNA was isolated using a pure link RNA mini kit (Invitrogen, Cat # 12183018A). Expression levels of PGC-1α, NRF-1, NRF-2, TFAM, and HO-1 mRNAs were determined using RT-qPCR. Primer pairs used for RT-PCR were as follows: PGC-1α 5’-GGC AGA AGG CAATTGAAG AG-3’ (F), 5’-TCA AAA CGGTCC CTC AGT TC-3’ (R), NRF-1 5’-CGCAAGTGGATCCTGACTGA-3’ (F), 5’-CCAATGTCACCACCTCCACA-3 (R), NRF-2 5’-CGGCTACGTTTCAGTCACTT-3’ (F), 5’-AGCTCCTCCCAAACTTGCTC-3’ (R), and TFAM 5’-CCG AGGTGG TTT TCATCTGT -3’ (F), 5’-GCATCT GGG TTC TGAGCT TT-3’ (R), HO-1 5’-TGGCATCTTCCCCAACGAAA-3’(F), 5’-ATGGCCGTGTCAACAAGGAT-’ (R) and the internal reference gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH): 5’-GGATTTGGTCGTATTGGG-3’ (F), 5’-GGAAGATGGTGATGGGATT-3’ (R) and hypoxanthine phosphoribosyltransferase (HPRT) 5’-GAAGAGCTATTGTAATGACC-3’ (F), 5’-GCGACCTTGACCATCTTTG-3’. Total RNA was isolated from the cells, and for reverse transcription reaction, 250 ng of RNA per sample was used in 20 µL reaction volumes. Besides, oligo (dTs) were also added to the reaction mixture. For this purpose, a cDNA synthesis kit from Invitrogen (Cat # 18080051) was used. Two control reactions—one without reverse transcriptase and the other without RNA—were also included for quality control. The reaction mixture for 20 µL comprised 9.5 µL DW, 2 µL MM1 (master mix, 1 µL of random hexamer, and 1 µL of dNTPs mix), 2.5 µL total RNA template (250 ng RNA), and then the tubes were incubated in a thermocycler (Aria Mxreal-time PCR system from Agilent, California, USA) for 5 min at 65°C followed by incubation on ice for 1 min. This was followed by the addition of 6 µL of MM2 (4 µL of 5X buffer, 1 µL of dithiothreitol, and 1 µL of superscript III reverse transcriptase enzyme) and incubated in the thermocycler for 5 min at 25°C, 50 min at 50°C and 15 min at 70°C. The final incubation was carried out for 15 min at 25°C and the tubes were then stored in a −20°C deep freezer. Protocol for the qPCR assay was the same as mentioned above for the determination of mtDNA to nDNA ratio, except for the internal references, which were GAPDH and HPRT in the case of gene expression study. Each sample was run in duplicate and the average Ct value was considered for the calculation of relative fold change. For delta Ct calculation geometric mean of the Ct values of internal reference genes, GAPDH and HPRT were used.
We performed a series of experiments; in experiment 1, the cells were incubated with four different doses of sitagliptin (10, 100, 1000, and 5000 µM), vildagliptin (10, 100, 1000, and 5000 µM), and exendin-4 (1, 10, and 100 nM). After completion of 72 h of incubation, media was removed and the cells were given a DPBS wash, followed by the addition of fresh media and further incubation for 48 h (Ligmusky et al., 2011; Choi et al., 2015; Iwaya et al., 2017; Sarkar et al., 2017; Yang et al., 2017; Zhang et al., 2018; Jang et al., 2019). After incubation, an MTT assay was performed. We also conducted a parallel setup in twelve well plates and acquired microscopic pictures to assess the apparent morphological changes in cells and in the cell number, if any. In experiment 2, we standardized the dose of doxorubicin at various concentrations (ranging from 1 to 10 µM). Among these doses, the one showing around 40% inhibition of cell viability (1 µM) was considered to clearly visualize the effect of incretin-based drugs. Based on these results, we conducted further experimentation considering the lowest effective dose of each drug which was 100 µM for both sitagliptin and vildagliptin and 10 nM for exendin-4. Furthermore, individual experiments were conducted to evaluate the influence of the above-mentioned drugs to assess viability, cell death, scratch assay, transwell migration, proliferation/morphological data, lactate concentration, mtDNA to nDNA ratio, gene expression analysis, and MitoTracker staining for mitochondrial membrane potential and biomass determination. Based on these data, we also examined the influence of sitagliptin, vildagliptin, and exendin-4 pre-treatment on the sensitivity of doxorubicin in TNBC cells through a viability assay. Subsequently, to determine the sensitivity of the drugs, we incubated the cells with 100 µM of sitagliptin and vildagliptin and 10 nM of exendin-4 for 72 h, then removed the media and replenished the cells with fresh medium. The cells were incubated with 1 µM of doxorubicin for 48 h, and then an MTT assay was performed to assess the changes in doxorubicin sensitivity after incretin-based pre-treatment.
All experimental data are represented as mean ± SEM. Statistical analysis was performed by one-way ANOVA (non-parametric), followed by post hoc Tukey’s multiple comparison test using Graph pad prism software 8.3 trial version. Significant difference was represented as *p < 0.05; **p < 0.01; ***p < 0.001 and ****p < 0.0001. Each experiment was repeated three times.
Dose standardization for sitagliptin, vildagliptin, and exendin-4 by MTT assay and morphological changes associated with loss of viability/apoptotic phenotype
In response to sitagliptin treatment at 10, 100, 1000, and 5000 μM, the percent (%) viability was 85.85, 70.46, 62.36, and 31.38, respectively (Fig. 1A), whereas in the case of vildagliptin, the respective percent viabilities were 94.17, 74.78, 59.35, and 51.92, as compared with the control group which was considered as 100% (Fig. 1B). Percent viability after treatment with exendin-4 at 1, 10, and 100 nM was 97.37, 75.15, and 63.34, respectively, compared with those of the control group (Fig. 1C). Similarly, in response to the treatment of sitagliptin, vildagliptin, and exendin-4 at all the above-mentioned doses for 72 h, apparent microscopic alterations in the cellular morphology and the reduction in cell number also revealed dose-dependent cytotoxic effects of these drugs on TNBC (Figs. 2A–2C). The untreated cells showed spindle-shaped morphology with a blurred outline, having a normal, healthy appearance with an adherent phenotype. On the other hand, cells treated with sitagliptin, vildagliptin, and exendin-4 showed round-shaped distorted morphology with detachment or poor adherence phenotype, suggesting cell death (Hu et al., 2016; Tripathi et al., 2022).
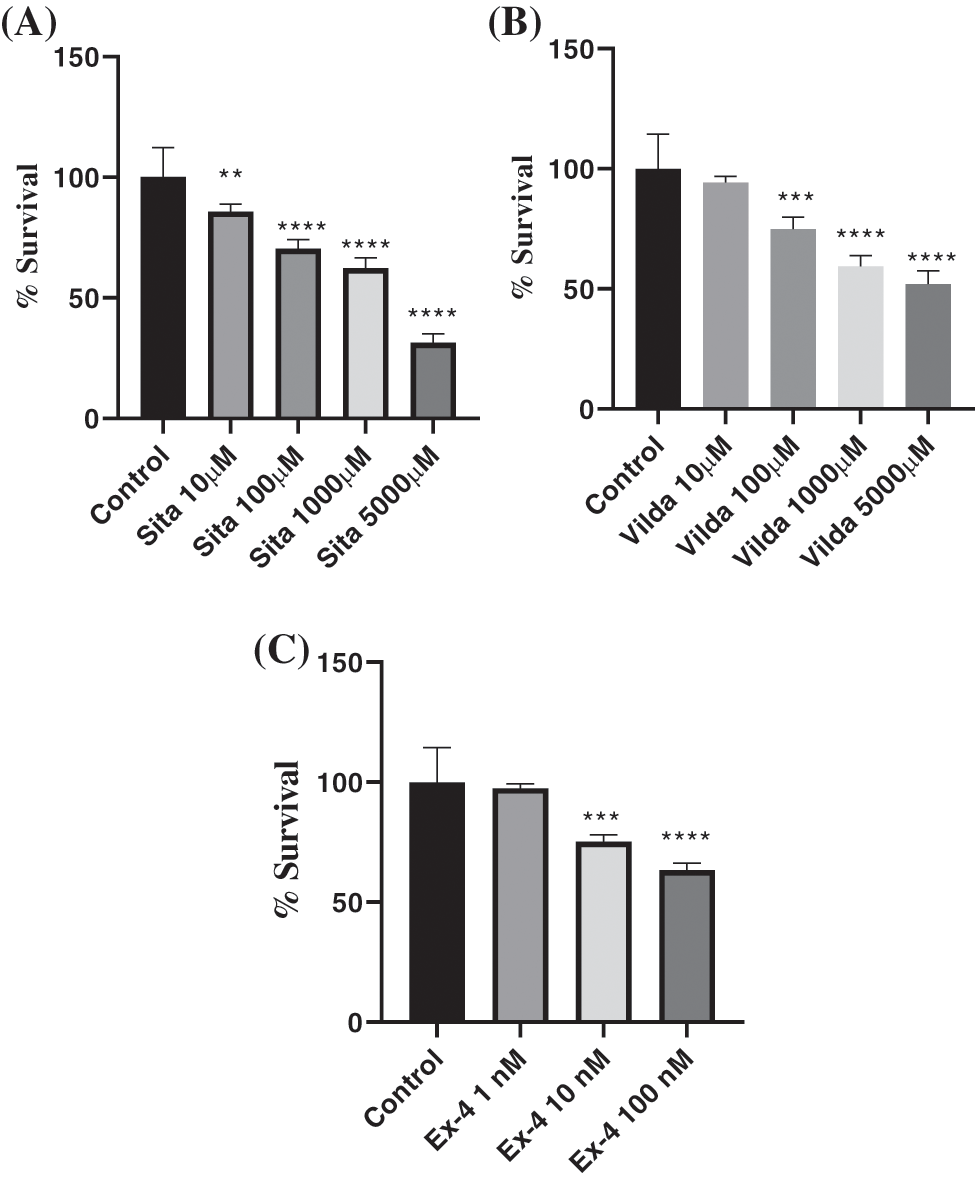
Figure 1: Effect of different doses of (A)sitagliptin, (B) vildagliptin, and (C) exendin-4 on the viability of MDA-MB-231 cells by using MTT assay. Data represented as mean ± SEM of n = 5 samples. Statistical significance was determined by using one-way ANOVA followed by post hoc Tukey’s multiple comparison test. Levels of significance were *p < 0.1; **p < 0.01; ***p < 0.001; ****p < 0.0001, as compared with the respective control groups.
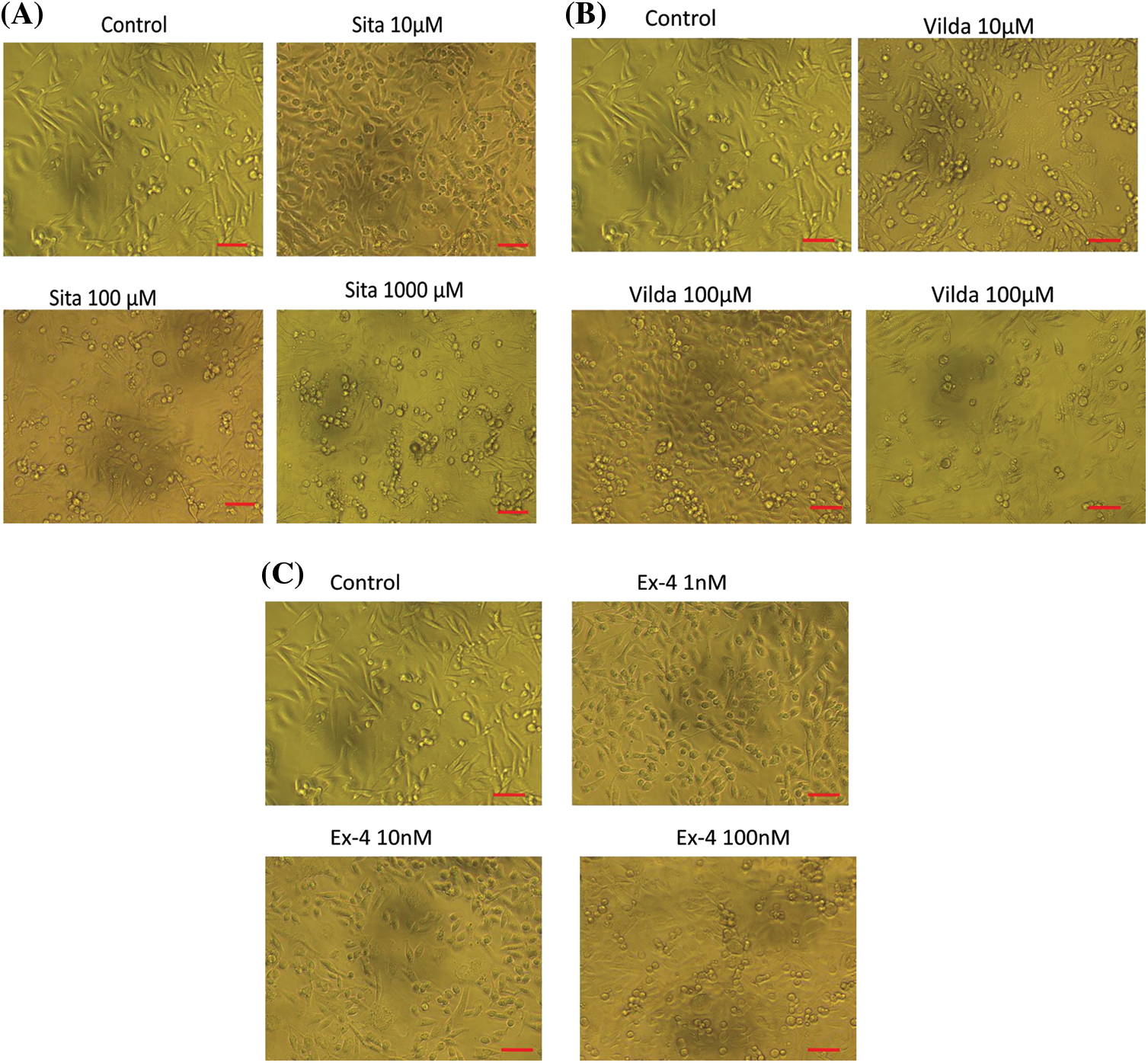
Figure 2: (A) Effect of sitagliptin, (B) vildagliptin, and (C) exendin-4on morphological alterations and decrease in the apparent number of cells. Cells were incubated in the absence and presence of all the studied doses of the drugs. After 72 h, apoptotic alterations in morphology were observed and imaged by an inverted light microscope (Scale bar, 100 µm).
The lowest possible dose was selected with 25% or more inhibition in MTT assay and a similar magnitude of the microscopic alterations in TNBC cells. Thus, selected doses of sitagliptin (100 μM), vildagliptin (100 μM), and exendin-4 (100 nM) were used for further experimentation.
Inhibition of TNBC cell migration (scratch and transwell migration assays)
Scratch assay showed that 72 h of sitagliptin, vildagliptin, and exendin-4 treatments decreased the cell migration (inhibition of wound closure) after 24 h by 46.51%, 44.92%, and 45.84% as compared with the control group (Figs. 3A and 3B). Similarly, transwell migration also showed a significant decrease in cell numbers per unit area migrated; transwell after pre-treatment with sitagliptin, vildagliptin, and exendin-4 transwell migration was 10.66, 14.50, and 9.30, respectively, as compared to the control wells, suggesting a significant inhibition of chemotactic migration (Figs. 3C and 3D).
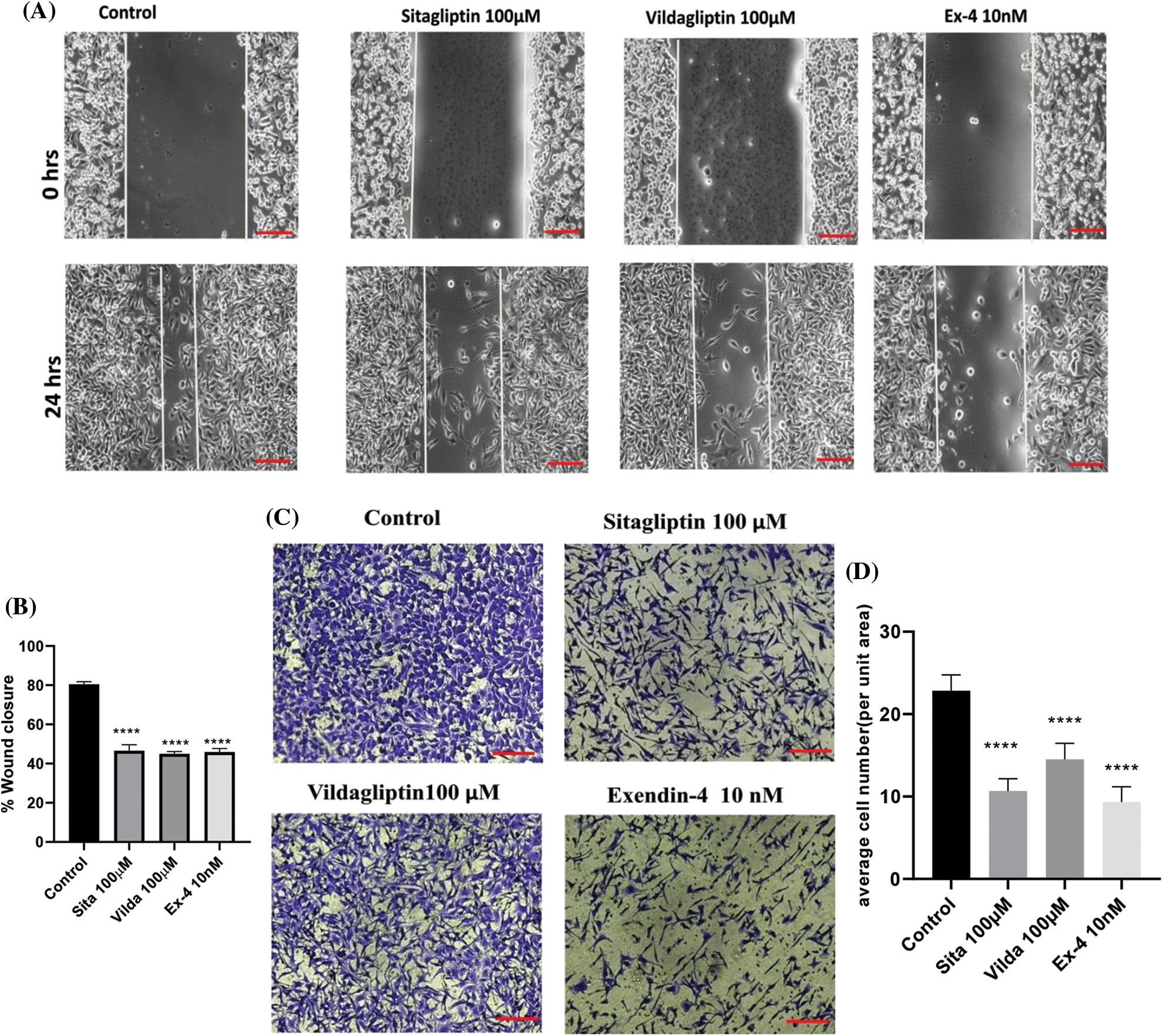
Figure 3: Effects of drugs (Sitagliptin, Vildagliptin & Exendin-4) on migratory potential of MDA-MB-231 by using scratch assay/wound healing and transwell migration. (A) Cells were incubated in the absence and presence of Sita (100 µM), Vilda (100 µM) and Exendin-4 (10 nM) and their effect was determined by percentage wound closure. Images were captured at 0 and 24 h after treatment. (B) Percentage wound closure was calculated by comparing the images of 0 and 24 h (scale bar 100 µm). (C) In transwell insert migration assay cells were treated with selected concentration of drugs for 72 h. Migratory cells in the lower surface of the insert were stained with crystal violet and imaged by an inverted light microscope. (D) The number of migratory cells was quantified by counting of cells per unit area of the image captured (Scale bar, 100 µm). Values are represented as mean ± SEM of n = 5 samples. Statistical significance was determined by one-way ANOVA followed by posthoc Tukey’s multiple comparison test. Level of significance was ***p < 0.001; ****p < 0.0001, as compared with control group.
ELISA showed a significant increase in the cell death of sitagliptin, vildagliptin, and exendin-4 treated cells compared to the control group. The percent increase in cell death was 33.61, 25.17, and 17.56, respectively, in sitagliptin, vildagliptin, and exendin-4 treated cells, as compared to the untreated control group (Fig. 4A).
In the conditioned media after pre-treatments with sitagliptin, vildagliptin, and exendin-4, there was a 30.25%, 25.21%, and 23.52% decrease respectively in the absorbance at 590 nm, compared with the control values (100%), consistently revealing a significant decrease in the amount of lactate formed (Fig. 4B).
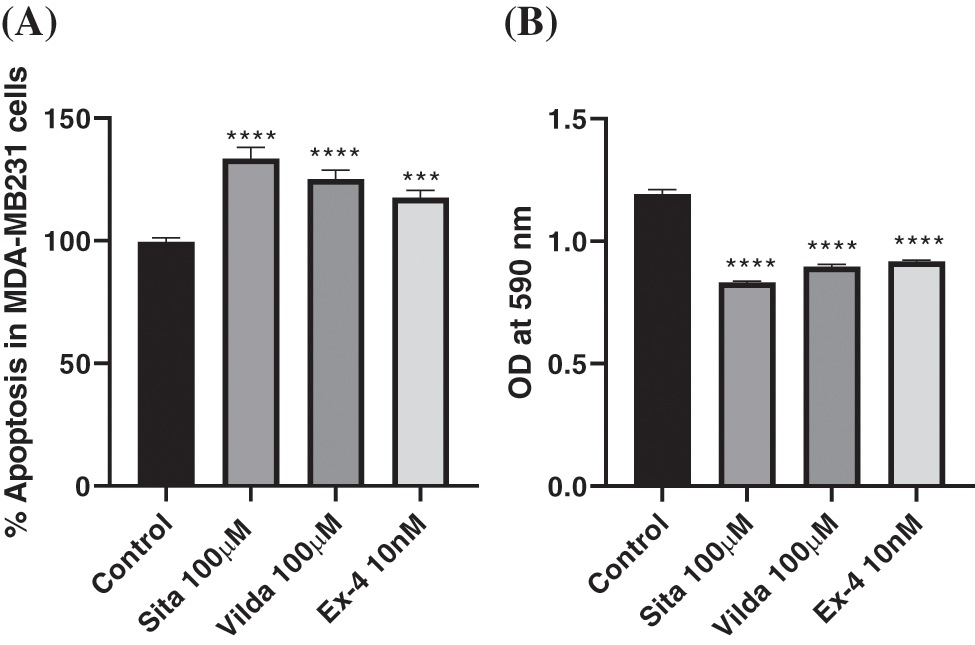
Figure 4: Effect of drugs (Sitagliptin, Vildagliptin & Exendin-4) on lactate concentration in conditioned media of MDA-MB-231 cells. (A) Percentage apoptosis was calculated as compared with the control group. (B) The amount of lactate produced was measured by taking the absorbance of the media at 590 nm. Values are represented as mean ± SEM of n = 3 samples. Statistical significance was determined by one-way ANOVA followed by posthoc Tukey’s multiple comparison test. Level of significance was ****p < 0.001; ****p < 0.0001, as compared with control group.
Mitochondrial DNA to nuclear DNA ratio
In response to pre-treatment with sitagliptin, vildagliptin, and exendin-4, the mt-DNA to n-DNA ratio increased by 3.564, 2.82, and 2.75-fold, respectively, compared with the control group (Fig. 5A). Data indicated an increase in mitochondrial biomass after treatment with sitagliptin, vildagliptin, and exendin-4.
Gene expression study of transcription factors involved in mitochondrial biogenesis
Analysis of gene expression by qPCR showed that in response to sitagliptin, vildagliptin, and exendin-4 treatment, there was an increase in the expression of transcription factors known to participate in mitochondrial biogenesis. Compared with the control group, after sitagliptin, vildagliptin, and exendin-4 treatments, respectively, relative fold increase in expressions of PGC-1α were 3.53, 2.53, and 2.13, NRF1 was 1.58, 1.40, and 1.49, NRF2 was 1.37, 1.26, and 1.37, TFAM was 1.93, 1.30, and 1.30, and finally, that of HO-1 was 1.42, 1.17, and 1.25 (Fig. 5B).
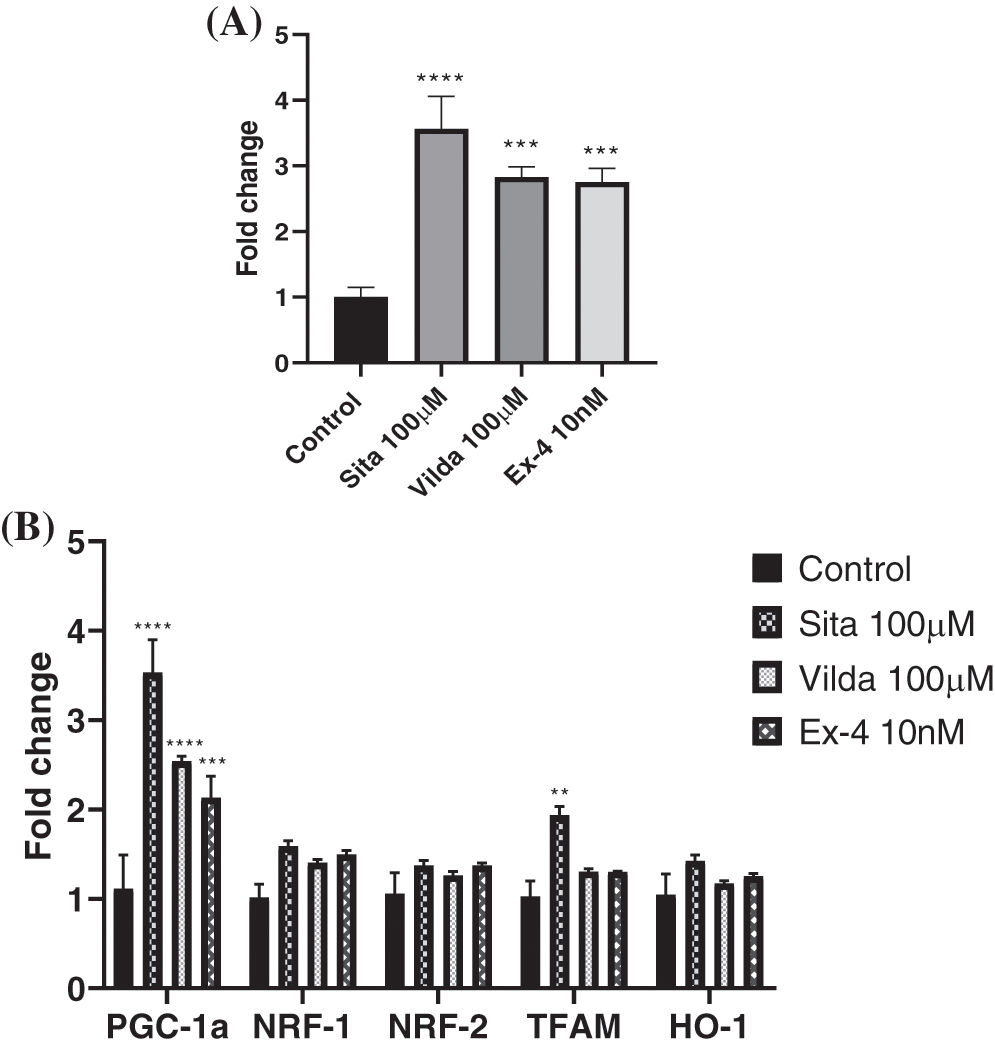
Figure 5: Effect of absence and presence of drugs (Sitagliptin, Vildagliptin & Exendin-4) for 72 h on mtDNA to nDNA ratio and on relative changes in the gene expression of mitochondrial biogenesis related genes in MDA-MB-231 cell line. (A) mtDNA to nDNA ratio was estimated by considering nDNA as a internal control and fold change was calculated as compared with the control group. (B) Relative fold change in NRF1, NRF2, PGC-1 alpha, TFAM and HO-1 compared to control group were studied. Housekeeping genes GAPDH and HPRT were considered as internal reference. Data represented as mean ± SEM of n = 3 samples and experiment was repeated three times. Statistical significance was determined by Two-way ANOVA followed by posthoc Tukey’s multiple comparison test Level of significance was *p < 0.05; ***p < 0.001; ****p < 0.0001, as compared to control group.
MitoTracker Green and MitoTracker Red staining
Treatment with sitagliptin, vildagliptin, and exendin-4 caused an increase in the fluorescence intensity of both MitoTracker Red and Green, revealing an increase in mitochondrial membrane potential as well as mitochondrial biogenesis. However, the increase in red fluorescence was moderate compared to green fluorescence in sitagliptin and vildagliptin-treated cells, suggesting a more pronounced increase in mitochondrial biomass than in membrane potential. (Figs. 6A and 6B).
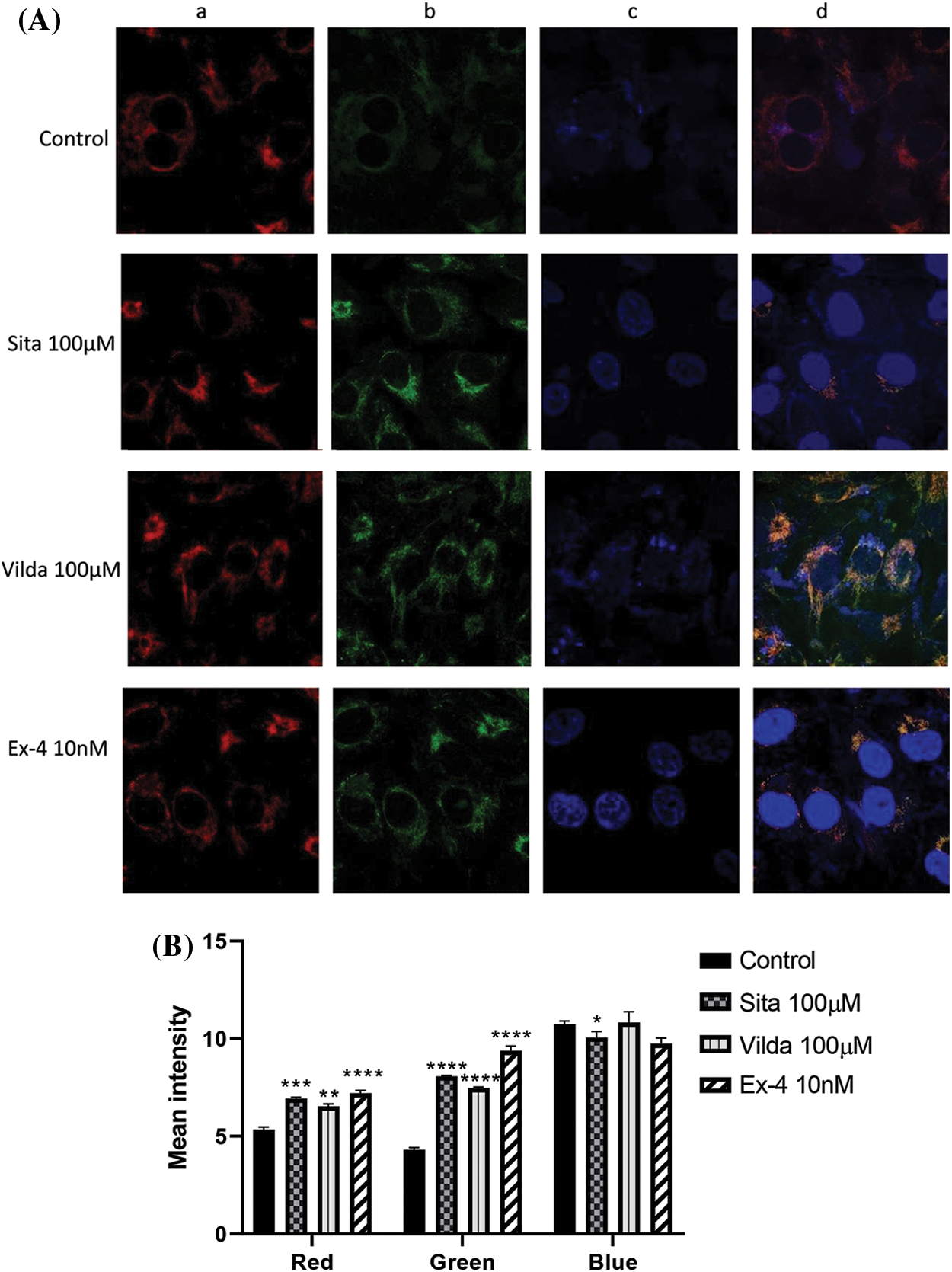
Figure 6: Effect of drugs (Sitagliptin, Vildagliptin & Exendin-4) on mitochondrial membrane potential & mitochondrial biomass. MDA-MB-231 cells were incubated in presence and absence of selected concentrations of drugs (Sitagliptin, Vildagliptin & Exendin-4) for 72 h. (A) To determine the difference in the mitochondrial membrane potential and mitochondrial biomass, treated and untreated cells were stained with Mitotracker RedFM & Mitotracker GreenFM, respectively. Nuclear staining was done by DAPI stain-(a) Image of Mitotracker RedFM stain, (b) Mitotracker GreenFM stain, (c) DAPI stain, (d) Merge image. (B) Fluorescence intensities were measured by confocal microscopy. Data represented as mean ± SEM of n = 3 samples. Statistical significance was determined by Two-way ANOVA followed by posthoc Tukey’s multiple comparison test. Level of significance was *p < 0.1; **p < 0.01; ***p < 0.001; ****p < 0.0001, as compared to control group.
Cytotoxicity evaluation of various doses of doxorubicin
MTT assay of MDA-MB-231 cells incubated with different doses of doxorubicin (ranging from 1 to 10 µM) for 48 h revealed a dose-dependent decrease in the viability of the cells. The highest reduction in viability of nearly 44.65% was observed at 10 µM, whereas the lowest viability was nearly 9.31% at 1 nM dose of doxorubicin. At 1 µM dose of doxorubicin, approximately 37.70% inhibition of viability was observed (Fig. 7A). Therefore, a dose of 1 µM of doxorubicin was considered to further evaluate the effect of test drugs on doxorubicin sensitivity.
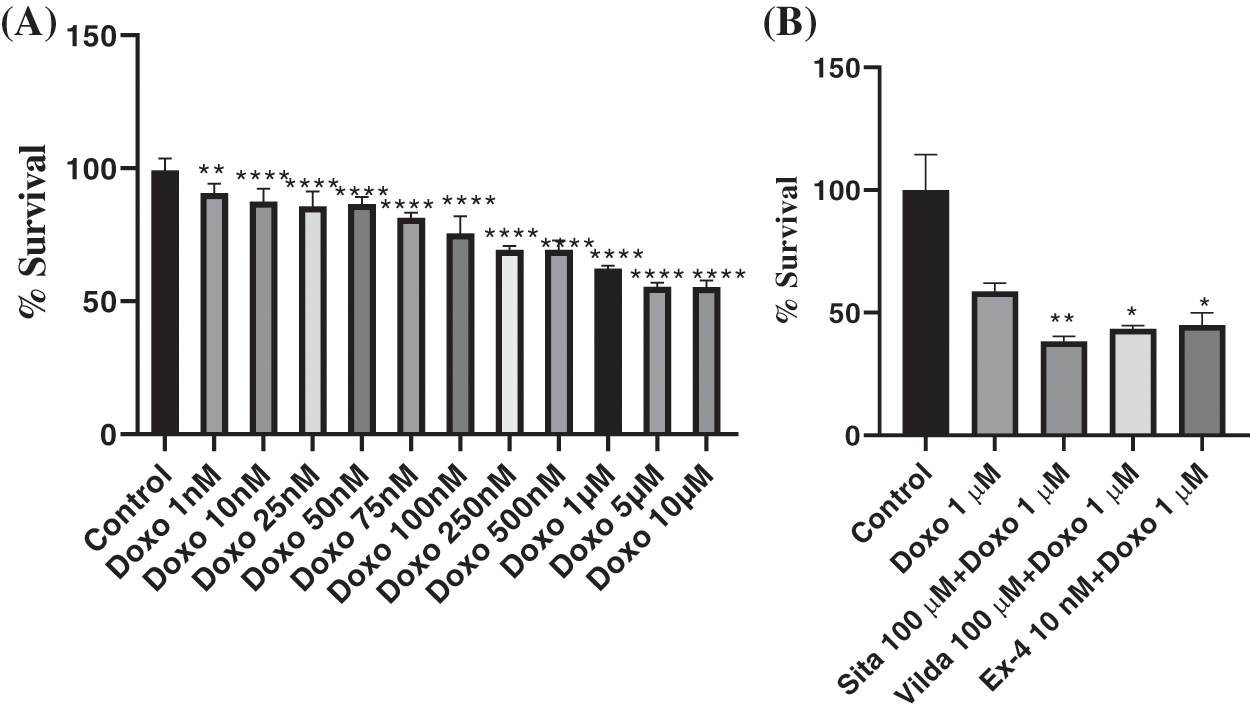
Figure 7: Effect of incretin based therapies on the sensitivity of doxorubicin treatment on MDA-MB-231 cell line was determined by MTT assay. (A) To determine the individual effect of doxorubicin on viability, cells were treated with various concentrations of doxorubicin (10 to 1 nM) for 48 h and then 1 μM (exerting nearly 40% inhibition on the viability of MDA-MB-231 cells) was selected for sensitivity experiment. (B) To determine the effect of sitagliptin, vildagliptin and exendin-4 at their selected dose on the sensitivity of doxorubicin, cells were incubated with these drugs for 72 h and then treated with 1 μM concentration of doxorubicin for 48 h. Data represented as mean ± SEM of n = 5 samples. Statistical significance was determined by using one-way ANOVA followed by posthoc Tukey’s multiple comparison test. Level of significance was *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001, as compared with the control group.
Pre-treatment of MDA-MB-231 cells with sitagliptin, vildagliptin, and exendin-4 at their respective selected dose for 72 h was followed by treatment with 1 µM doxorubicin for 48 h in fresh media. MTT assay revealed that singly, doxorubicin treatment inhibited the cell viability by 41.24%, whereas pre-treatment with sitagliptin, vildagliptin, and exendin-4 followed by doxorubicin treatment inhibited cell viability by 61.68%, 56.52%, and 55.01%, respectively. These data clearly revealed an increase in doxorubicin sensitivity after pre-treatment with these incretin-based drugs (Fig. 7B).
Role of DPP-IV in the progression of metastatic breast cancer is documented by various studies; accordingly, a number of reports have suggested anti-cancer effects of GLP-1 agonists and DPP-IV inhibitors (Jaiswal et al., 2022). In the present study, we observed anti-cancer effects of DPP-IV inhibitors, sitagliptin, and vildagliptin, wherein the drugs decreased the viability, proliferation, and migratory potential of MDA-MB-231 cells along with the induction of apoptosis, which might be mediated through inhibition of DPP-IV, as MDA-MB-231 are TNBC cells that express DPP-IV (Pascual Alonso et al., 2022). Dose-dependent effects of exendin-4 (GLP-1R agonist) showed that the highest loss in viability was observed at 100 nM, whereas minimum loss of viability was observed at 1 nM. These data are in agreement with the earlier studies, which reported that low doses could inhibit viability or proliferation but could not induce apoptosis, which is possibly due to the difference in the magnitude of the inhibition of pro-survival Akt and inflammatory NF-kB pathways (Jaiswal et al., 2022). Similarly, a dose-dependent decrease in cell viability by sitagliptin and vildagliptin might be the result of the magnitude of DPP-IV inhibition in dose dependent manner which is known to participate in the process of carcinogenesis through various mechanisms (Jaiswal et al., 2022). DPP-IV induces EGF expression, which in turn leads to the activation of PIN1; so, on the one hand, increase in EGF level promotes MEK/ERK and JNK/c-Jun signaling and AP-1 activity which ultimately leads to the cell viability, growth and cell transformation, on the other hand, PIN1 is a peptidylprolyl cis/trans isomerase which is a novel phosphorylation signaling regulator controlling the proliferation and transformation (Choi et al., 2015; Lu and Zhou, 2007). Cis/trans isomerization of Ser-Pro or Thr-Pro peptide bonds targets several proteins that alter the stability, localization, and protein-protein interactions (Liou et al., 2011), and these conformational changes have profound effects on targets proteins such as p53, cyclinD1, c-Jun, MAPK kinase1, NF-kB, and STAT3, that influence cell cycle progression and differentiation decisions (Jaiswal et al., 2022; Wulf et al., 2001; Zheng et al., 2002; Ryo et al., 2003; Fukuchi et al., 2006; Khanal et al., 2010; Kim et al., 2014). Therefore, sitagliptin and vildagliptin treatment might have reversed the carcinogenic effects of the DPP-IV. Interestingly, MDA-MB-231 cells exhibit Warburg metabolism, but in response to sitagliptin and vildagliptin treatments, we also observed increased mitochondrial biogenesis as revealed by the increase in MitoTracker staining, mtDNA to nDNA ratio, and increased expression of the transcription factors involved in mitochondrial biogenesis along with the decrease in lactate level in conditioned media, suggesting switching of Warburg metabolism into anti-Warburg effect and normalization of the cancer phenotype of the cells. Other reports have further suggested a decrease in mitochondrial number; besides, mutations in mtDNA have been correlated with the tumor progression and poor prognosis of breast cancer (Tseng et al., 2006; Yu et al., 2007). Mitochondrial metabolism is a double edge sword in treating cancer cells, as Warburg metabolism or poor mitochondrial functioning is being found to be associated with resistance to apoptosis, increased genome instability, chemo-resistance, proliferation, invasion, migration, and increased stemness of the cancer cells (Gogvadze et al., 2009; Seyfried and Shelton, 2010; Indran et al., 2011; Bhandary et al., 2012; Ward and Thompson, 2012; Wallace, 2012; Han et al., 2013; Loureiro et al., 2013; Sciacovelli et al., 2014; Gaude and Frezza, 2014; Zong et al., 2016; Liu et al., 2018), whereas mitochondria are also essential for cancer cell proliferation to resist oxidative stress and reliance of cancer cells on oxidative phosphorylation for metastasis by PGC-1α-mediated mitochondrial biogenesis (Le Bleu et al., 2014; Tan et al., 2015; Tan et al., 2017; Paliwal et al., 2021; Faubert et al., 2020). Contrasting reports on mitochondrial functioning and its association with cancer progression suggest that while a modest improvement in mitochondrial metabolism through mitochondrial biogenesis or improvements in their functioning may diminish the process of carcinogenesis and severity of cancer, the same outcome may be achieved by severe depletion of mitochondrial functioning. In the present study, the anti-cancer effects of incretin-based drugs on TNBC seem to be an outcome of the modest increase in mitochondrial biogenesis and/or improved mitochondrial functioning. In response to the treatment of sitagliptin, vildagliptin, and exendin-4, an increase in apoptosis and increased chemosensitivity of doxorubicin are also results of modest improvement in mitochondrial biogenesis, as the decrease in mitochondrial membrane potential, mitochondrial biomass and deletion of mitochondrial DNA are associated with anti-apoptotic phenotype, as both extrinsic and intrinsic pathways of apoptosis depend upon the mitochondria (Indran et al., 2011; Shen et al., 2018). The generation of reactive oxygen species in the mitochondria of cancer cells also leads to mutations in the mitochondrial genome and ultimately compromises its function. Metabolism-directed mutations have also been recently revealed to participate in the progression of cancer (Ward and Thompson, 2012; Kamp et al., 2016), which further consolidate the results of the present study. Theobserved anti-cancer effects of exendin-4 are also in line with the earlier reports that have shown to inhibit growth, migration, invasion, and apoptotic induction in breast and ovarian cancer cells (Fidan-Yaylalı et al., 2016; He et al., 2016). These effects were suggested as a result of the inhibition of the PI3/AKT pathway (Fidan-Yaylalı et al., 2016). Besides, upregulation of E-cadherin was thought to be involved in reducing migration and invasion of tumor cells and act as a strong tumor suppressor in cancer progression and pro-apoptotic phenotype through induction of cleaved caspase-3 expression (Nie et al., 2018). In response to exendin-4 treatment, we also observed a modest increase in the mtDNA to nDNA ratio, MitoTracker staining, and the expression of transcription factors involved in mitochondrial biogenesis PGC-1alpha, NRF-1, NRF-2, and TFAM, along with a decrease in lactate concentration in conditioned media, suggesting improvements in the mitochondrial functioning in the cancer cells that reversed the Warburg metabolism up to an extent through which normalization of the cancer phenotype might have occurred. Importantly, lactic acid has been correlated well with the migratory potential of cancer cells and metastatic phenotype, as lactic acid activates the vascular endothelial growth factor (VEGF), TGF-β, IL-1, and HIF-α and these participate in promoting invasion and migration of cancer cells (Jiang, 2017). These findings further support our data that the decrease in migration of TNBC cells in response to sitagliptin, vildagliptin, and exendin-4 might be mediated via a decrease in a glycolytic shift which is regulated by HIF-α and higher lactic acid feedback to further enhance the glycolysis in cancer cells (Jiang, 2017).
Increased doxorubicin chemo-sensitivity after pre-treatment with exendin-4 is another important outcome which is also in line with the earlier reports suggesting that anti-Warburg rearrangements support chemotherapeutic response in breast cancer (Suhane and Ramanujan, 2011).
Therefore, based on the data from the present study, we conclude that sitagliptin, vildagliptin, and exendin-4 have anti-cancer effects and improve the mitochondrial functioning via the increase in mitochondrial biogenesis may be a major underlying mechanism. Finally, the magnitude of anti-cancer effects was in the order of: sitagliptin > vildagliptin > exendin-4.
Acknowledgement: We acknowledge the facilities provided by NCCS, Pune and the School of Biotechnology, DAVV for our experimental work.
Availability of Data and Materials: All data generated or analyzed during this study are included in this article.
Author Contribution: The authors confirm contribution to the paper is as follows: study conception and design: Hamendra Singh Parmar; data collection: Pooja Jaiswal, Versha Tripathi; analysis and interpretation of results: Pooja Jaiswal, Versha Tripathi, Anshul Assaiya, Dharmendra Kashyap, Hamendra Singh Parmar; draft manuscript preparation: Janesh Kumar, Hem Chandra Jha, Rajesh Sharma, Amit Kumar Dixit, Rahul Dubey, Anamika Singh, Hamendra Singh Parmar. All authors reviewed the results and approved the final version of the manuscript.
Ethics Approval: Not applicable.
Funding Statement:The work was supported by the Council of Scientific and Industrial Research-Junior Research Fellowship to Ms. Pooja Jaiswal (File No. 09/301 (0137)/2019-EMR-I) and Ms. Versha Tripathi (09/301 (0138)/2019-EMR-I). We also acknowledge the India-Belarus joint project (DST/INT/BLR/P-24/2019) funded by the Department of Science and Technology, New Delhi and the research grant from Devi Ahilya University, Indore (Dev/Seedmoney2.0/2020-21/655) sanctioned to Dr. Hamendra Singh Parmar.
Conflicts of Interest:The authors declare that they have no conflicts of interest to report regarding the present study.
References
Abuelezz SA, Hendawy N, Abdel Gawad S (2016). Alleviation of renal mitochondrial dysfunction and apoptosis underlies the protective effect of sitagliptin in gentamicin-induced nephrotoxicity. Journal of Pharmacy and Pharmacology 68: 523–532. DOI 10.1111/jphp.12534. [Google Scholar] [CrossRef]
Apaijai N, Pintana H, Chattipakorn SC, Chattipakorn N (2013). Effects of vildagliptin versus sitagliptin, on cardiac function, heart rate variability and mitochondrial function in obese insulin-resistant rats. British Journal of Pharmacology 169: 1048–1057. DOI 10.1111/bph.12176. [Google Scholar] [CrossRef]
Avagliano A, Ruocco MR, Aliotta F, Belviso I, Accurso A et al. (2019). Mitochondrial flexibility of breast cancers: A growth advantage and a therapeutic opportunity. Cells 8: 401. DOI 10.3390/cells8050401. [Google Scholar] [CrossRef]
Ayyasamy V, Owens KM, Desouki MM, Liang P, Bakin A et al. (2011). Cellular model of Warburg effect identifies tumor promoting function of UCP2 in breast cancer and its suppression by genipin. PLoS One 6: e24792. DOI 10.1371/journal.pone.0024792. [Google Scholar] [CrossRef]
Bhandary B, Marahatta A, Kim HR, Chae HJ (2012). Mitochondria in relation to cancer metastasis. Journal of Bioenergetics and Biomembranes 44: 623–627. DOI 10.1007/s10863-012-9464-x. [Google Scholar] [CrossRef]
Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D et al. (2006). Race, breast cancer subtypes, and survival in the carolina breast cancer Study. Journal of the American Medical Association 295: 2492–2502. DOI 10.1001/jama.295.21.2492. [Google Scholar] [CrossRef]
Choi HJ, Kim JY, Lim SC, Kim G, Yun HJ et al. (2015). Dipeptidyl peptidase 4 promotes epithelial cell transformation and breast tumourigenesis via induction of PIN1 gene expression. British Journal of Pharmacology 172: 5096–5109. DOI 10.1111/bph.13274. [Google Scholar] [CrossRef]
Fadaka A, Ajiboye B, Ojo O, Adewale O, Olayide I et al. (2017). Biology of glucose metabolization in cancer cells. Journal of Oncological Science 3: 45–51. DOI 10.1016/j.jons.2017.06.002. [Google Scholar] [CrossRef]
Faubert B, Solmonson A, de Berardinis RJ (2020). Metabolic reprogramming and cancer progression. Science 368: eaaw 5473. DOI 10.1126/science.aaw5473. [Google Scholar] [CrossRef]
Feng W, Gentles A, Nair RV, Huang M, Lin Y et al. (2014). Targeting unique metabolic properties of breast tumor initiating cells. Stem Cells 32: 1734–1745. DOI 10.1002/stem.1662. [Google Scholar] [CrossRef]
Fidan-Yaylalı G, Dodurga Y, Seçme M, Elmas L (2016). Antidiabetic exendin-4 activates apoptotic pathway and inhibits growth of breast cancer cells. Tumour Biology 37: 2647–2653. DOI 10.1007/s13277-015-4104-9. [Google Scholar] [CrossRef]
Fukuchi M, Fukai Y, Kimura H, Sohda M, Miyazaki T et al. (2006). Prolyl isomerase Pin1 expression predicts prognosis in patients with esophageal squamous cell carcinoma and correlates with cyclinD1 expression. International Journal of Oncology 29: 329–334. DOI 10.3892/ijo.29.2.329. [Google Scholar] [CrossRef]
Gaude E, Frezza C (2014). Defects in mitochondrial metabolism and cancer. Cancer Metabolism 2: 10. DOI 10.1186/2049-3002-2-10. [Google Scholar] [CrossRef]
Gogvadze V, Orrenius S, Zhivotovsky B (2009). Mitochondria as targets for cancer chemotherapy. Seminars in Cancer Biology 19: 57–66. DOI 10.1016/j.semcancer.2008.11.007. [Google Scholar] [CrossRef]
Han T, Kang D, Ji D, Wang X, Zhan W (2013). How does cancer cell metabolism affect tumor migration and invasion? Cell Adhesion and Migration 7: 395–403. DOI 10.4161/cam.26345. [Google Scholar] [CrossRef]
He W, Yu S, Wang L, He M, Cao X et al. (2016). Exendin-4 inhibits growth and augments apoptosis of ovarian cancer cells. Molecular and Cellular Endocrinolgy 436: 240–249. DOI 10.1016/j.mce.2016.07.032. [Google Scholar] [CrossRef]
Hu H, Dong Z, Tan P, Zhang Y, Liu L et al. (2016). Antibiotic drug tigecycline inhibits melanoma progression and metastasis in a p21CIP1/Waf1-dependent manner. Oncotarget 7: 3171–3185. DOI 10.18632/oncotarget.6419. [Google Scholar] [CrossRef]
Igoillo-Esteve M, Oliveira AF, Cosentino C, Fantuzzi F, Demarez C et al. (2020). Exenatide induces frataxin expression and improves mitochondrial function in Friedreich ataxia. Journal of Clinical Investigation (Insight) 5: e134221. DOI 10.1172/jci.insight.134221. [Google Scholar] [CrossRef]
Indran IR, Tufo G, Pervaiz S, Brenner C (2011). Recent advances in apoptosis, mitochondria and drug resistance in cancer cells. Biochimica et Biophysicsa Acta 1807: 735–745. DOI 10.1016/j.bbabio.2011.03.010. [Google Scholar] [CrossRef]
Iwaya C, Nomiyama T, Komatsu S, Kawanami T, Tsutsumi Y et al. (2017). Exendin-4, a Glucagon like peptide-1 receptor agonist, attenuates breast cancer growth by inhibiting NF-kB activation. Endocrinology 158: 4218–4232. DOI 10.1210/en.2017-00461. [Google Scholar] [CrossRef]
Jaiswal P, Tripathi V, Nayak A, Kataria S, Lukashevich V et al. (2022). A molecular link between diabetes and breast cancer: Therapeutic potential of repurposing incretin-based therapies for breast cancer. Current Cancer Drug Targets 21: 829–848. DOI 10.2174/1568009621666210901101851. [Google Scholar] [CrossRef]
Jang JH, Janker F, Meester ID, Yamada Y, Limani P et al. (2019). Suppression of lung metastases by the CD26/DPP4 inhibitor Vildagliptin in mice. Clinical and Experimental Metastasis 32: 677–687. DOI 10.1007/s10585-015-9736-z. [Google Scholar] [CrossRef]
Jiang B (2017). Aerobic glycolysis and high level of lactate in cancer metabolism and microenvironment. Genes and Diseases 4: 25–27. DOI 10.1016/j.gendis.2017.02.003. [Google Scholar] [CrossRef]
Kamp WM, Wang PY, Hwang PM (2016). TP53 mutation, mitochondria and cancer. Current Opinion in Genetics and Development 38: 16–22. DOI 10.1016/j.gde.2016.02.007. [Google Scholar] [CrossRef]
Kang MY, Oh TJ, Cho YM (2015). Glucagon-like peptide-1 increases mitochondrial biogenesis and function in INS-1 rat insulinoma cells. Endocrinology and Metabolism 30: 216–220. DOI 10.3803/EnM.2015.30.2.216. [Google Scholar] [CrossRef]
Khanal P, Namgoong GM, Kang BS, Woo ER, Choi HS (2010). The prolylisomerase Pin1 enhances HER-2 expression and cellular transformation via its interaction with mitogen-activated protein kinase/extracellular signal-regulated kinase kinase 1. Molecular Cancer Therapeutics 9: 606–616. DOI 10.1158/1535-7163.MCT-09-0560. [Google Scholar] [CrossRef]
Kim K, Kim G, Kim JY, Yun HJ, Lim SC et al. (2014). Interleukin-22 promotes epithelial cell transformation and breast tumorigenesis via MAP3K8 activation. Carcinogenesis 35: 1352–1361. DOI 10.1093/carcin/bgu044. [Google Scholar] [CrossRef]
Kim S, Kimdo H, Jung WH, Koo JS (2013). Metabolic phenotypes in triple-negative breast cancer. Tumour Biology 34: 1699–1712. DOI 10.1007/s13277-013-0707-1. [Google Scholar] [CrossRef]
Le Bleu VS, O’Connell JT, Gonzalez Herrera KN, Wikman H, Pantel K et al. (2014). PGC-1α mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nature Cell Biology 16: 992–1003. DOI 10.1038/ncb3039. [Google Scholar] [CrossRef]
Li Y, Glotfelty EJ, Karlsson T, Fortuno LV, Harvey BK et al. (2021). The metabolite GLP-1 (9-36) is neuroprotective and anti-inflammatory in cellular models of neurodegeneration. Journal of Neurochemistry 159: 867–886. DOI 10.1111/jnc.15521. [Google Scholar] [CrossRef]
Ligmusky H, Ido W, Shira I, Michal H, Sarah F et al. (2011). The peptide-hormone glucagon-like peptide-1 activates cAMP and inhibits growth of breast cancer cells. Breast Cancer Research and Treatment 132: 449–461. DOI 10.1007/s10549-011-1585-0. [Google Scholar] [CrossRef]
Liou YC, Zhou XZ, Lu KP (2011). Prolylisomerase Pin1 as a molecular switch to determine the fate of phosphoproteins. Trends in Biochemical Sciences 36: 501–514. DOI 10.1016/j.tibs.2011.07.001. [Google Scholar] [CrossRef]
Liu Y, Tong L, Luo Y, Li X, Chen G et al. (2018). Resveratrol inhibits the proliferation and induces the apoptosis in ovarian cancer cells via inhibiting glycolysis and targeting AMPK/mTOR signaling pathway. Journal of Cellular Biochemistry 119: 6162–6172. DOI 10.1002/jcb.26822. [Google Scholar] [CrossRef]
Loureiro R, Mesquita KA, Oliveira PJ, Vega-Naredo I (2013). Mitochondria in cancer stem cells: A target for therapy. Recent Patents on Endocrine, Metabolic & Immune Drug Discovery 7: 102–114. DOI 10.2174/18722148113079990006. [Google Scholar] [CrossRef]
Lu KP, Zhou XZ (2007). The prolylisomerase PIN1: A pivotal new twist in phosphorylation signalling and disease. Nature Reviews Molecular Cell Biology 8: 904–916. DOI 10.1038/nrm2261. [Google Scholar] [CrossRef]
Mostafa Tork O, Ahmed Rashed L, BakrSadek N, Abdel-Tawab MS (2019). Targeting altered mitochondrial biogenesis in the brain of diabetic rats: Potential effect of pioglitazone and exendin-4. Reports of Biochemistry and Molecular Biology 8: 287–300. [Google Scholar]
Nasser MI, Masood M, Wei W, Li X, Zhou Y et al. (2017). Cordycepin induces apoptosis in SGC7901 cells through mitochondrial extrinsic phosphorylation of PI3K/Akt by generating ROS. International Journal of Oncology 50: 911–919. DOI 10.3892/ijo.2017.3862. [Google Scholar] [CrossRef]
Nath M, Bhattacharjee K, Choudhury Y (2021). Vildagliptin, a dipeptidyl peptidase-4 inhibitor, reduces betel-nut induced carcinogenesis in female mice. Life Science 266: 118870. DOI 10.1016/j.lfs.2020.118870. [Google Scholar] [CrossRef]
Nie ZJ, Zhang YG, Chang YH, Li QY, Zhang YL (2018). Exendin-4 inhibits glioma cell migration, invasion and epithelial-to-mesenchymal transition through GLP-1R/sirt3 pathway. Biomedical Pharmacotherapy 106: 1364–1369. DOI 10.1016/j.biopha.2018.07.092. [Google Scholar] [CrossRef]
Paliwal S, Chaudhuri R, Agrawal A, Mohanty S (2021). Regenerative abilities of mesenchymal stem cells through mitochondrial transfer. Journal of Biomedical Science 25: 31. DOI 10.1186/s12929-018-0429-1. [Google Scholar] [CrossRef]
Parmar HS, Nayak A, Gavel PK, Jha HC, Bhagwat S et al. (2021). Cross talk between COVID-19 and breast cancer. Current Cancer Drug Targets 21: 575–600. DOI 10.2174/1568009621666210216102236. [Google Scholar] [CrossRef]
Parmar HS, Nayak A, Kataria S, Tripathi V, Jaiswal P et al. (2022). Restructuring the ONYX-015 adenovirus by using spike protein genes from SARS-CoV-2 and MERS-CoV: Possible implications in breast cancer treatment. Medical Hypotheses 159: 110750. DOI 10.1016/j.mehy.2021.110750. [Google Scholar] [CrossRef]
Pascual Alonso I, Valiente PA, Valdes-Tresanco ME, Arrebola Y, Almeida Garcia F et al. (2022). Discovery of tight-binding competitive inhibitors of dipeptidyl peptidase IV. International Journal of Biological Macromolecules 196: 120–130. DOI 10.1016/j.ijbiomac.2021.12.056. [Google Scholar] [CrossRef]
Rouzier R, Perou CM, Symmans WF, Ibrahim N, Cristofanilli M et al. (2005). Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clinical Cancer Research 11: 5678–5685. DOI 10.1158/1078-0432.CCR-04-2421. [Google Scholar] [CrossRef]
Ryo A, Suizu F, Yoshida Y, Perrem K, Liou YC et al. (2003). Regulation of NF-kappaB signaling by Pin1-dependent prolyl isomerization and ubiquitin-mediated proteolysis of p65/RelA. Molecular Cell 12: 1413–1426. DOI 10.1016/S1097-2765(03)00490-8. [Google Scholar] [CrossRef]
Sarkar M, Dey S, Giri B (2017). Antiproliferative and apoptogenic efficacy of antidiabetic drugs metformin and sitagliptin against MCF7 and HEPG2 cancer cells: A comparative molecular study. Journal of Drug Delivery and Therapeutics 7: 11–21. DOI 10.22270/jddt.v7i6.1519. [Google Scholar] [CrossRef]
Sciacovelli M, Gaude E, Hilvo M, Frezza C (2014). The metabolic alterations of cancer cells. Methods in Enzymology 542: 1–23. DOI 10.1016/B978-0-12-416618-9.00001-7. [Google Scholar] [CrossRef]
Seyfried TN, Shelton LM (2010). Cancer as a metabolic disease. Nutrition and Metabolism 7: 7. DOI 10.1186/1743-7075-7-7. [Google Scholar] [CrossRef]
Shen X, Gao X, Li H, Gu Y, Wang J (2018). TIMP-3 increases the chemosensitivity of laryngeal carcinoma to cisplatin via facilitating mitochondria-dependent apoptosis. Oncology Research 27: 73–80. DOI 10.3727/096504018X15201099883047. [Google Scholar] [CrossRef]
Strycharz J, Rygielska Z, Swiderska E, Drzewoski J, Szemraj J et al. (2018). SIRT1 as a therapeutic target in diabetic complications. Current Medical Chemistry 25: 1002–1035. DOI 10.2174/0929867324666171107103114. [Google Scholar] [CrossRef]
Suhane S, Ramanujan VK (2011). Thyroid hormone differentially modulates Warburg phenotype in breast cancer cells. Biochemical and Biophysical Research Communications 414: 73–78. DOI 10.1016/j.bbrc.2011.09.024. [Google Scholar] [CrossRef]
Tan AS, Baty JW, Dong LF, Bezawork-Geleta A, Endaya B et al. (2015). Mitochondrial genome acquisition restores respiratory function and tumorigenic potential of cancer cells without mitochondrial DNA. Cell Metabolism 21: 81–94. DOI 10.1016/j.cmet.2014.12.003. [Google Scholar] [CrossRef]
Tan J, Song M, Zhou M, Hu Y (2017). Antibiotic tigecycline enhances cisplatin activity against human hepatocellular carcinoma through inducing mitochondrial dysfunction and oxidative damage. Biochemical and Biophysical Research Communications 483: 17–23. DOI 10.1016/j.bbrc.2017.01.021. [Google Scholar] [CrossRef]
Tang X, Lin CC, Spasojevic I, Iversen ES, Chi JT et al. (2014). A joint analysis of metabolomics and genetics of breast cancer. Breast Cancer Research 16: 1–15. DOI 10.1186/s13058-014-0415-9. [Google Scholar] [CrossRef]
Tripathi V, Jaiswal P, Assaiya A, Kumar J, Parmar HS (2022). Anti-cancer effects of AICAR on triple-negative breast cancer (TNBC) cells: Mitochondrial modulation may be an underlying mechanism. Current Cancer Drug Targets (in Press). [Google Scholar]
Tseng LM, Yin PH, Chi CW, Hsu CY, Wu CW et al. (2006). Mitochondrial DNA mutations and mitochondrial DNA depletion in breast cancer. Genes Chromosome and Cancer 45: 629–638. DOI 10.1002/(ISSN)1098-2264. [Google Scholar] [CrossRef]
Wallace DC (2012). Mitochondria and cancer. Nature Reviews Cancer 12: 685–698. DOI 10.1038/nrc3365. [Google Scholar] [CrossRef]
Ward PS, Thompson CB (2012). Metabolic reprogramming: A cancer hallmark even Warburg did not anticipate. Cancer Cell 21: 297–308. DOI 10.1016/j.ccr.2012.02.014. [Google Scholar] [CrossRef]
Weng G, Zhou B, Liu T, Huang Z, Yang H (2019). Sitagliptin promotes mitochondrial biogenesis in human SH-SY5Y cells by increasing the expression of PGC-1α/NRF1/TFAM. IUBMB Life 71: 1515–1521. DOI 10.1002/iub.2076. [Google Scholar] [CrossRef]
Wulf GM, Ryo A, Wulf GG, Lee SW, Niu T et al. (2001). Pin1 is overexpressed in breast cancer and cooperates with Ras signaling in increasing the transcriptional activity of c-Jun towards cyclin D1. Journal of European Molecular Biology Organization 20: 3459–3472. DOI 10.1093/emboj/20.13.3459. [Google Scholar] [CrossRef]
Yang X, Zhang X, Wu R, Huang Q, Jiang Y et al. (2017). DPPIV promotes endometrial carcinoma cell proliferation, invasion and tumorigenesis. Oncotarget 8: 8679–8692. DOI 10.18632/oncotarget.14412. [Google Scholar] [CrossRef]
Yang Q, Ai W, Nie L, Yan C, Wu S (2020). Vildagliptin reduces myocardial ischemia-induced arrhythmogenesis via modulating inflammatory responses and promoting expression of genes regulating mitochondrial biogenesis in rats with type-II diabetes. Journal of Interventional Cardiac Electrophysiology 59: 517–526. DOI 10.1007/s10840-019-00679-9. [Google Scholar] [CrossRef]
Yu M, Zhou Y, Shi Y, Ning L, Yang Y et al. (2007). Reduced mitochondrial DNA copy number is correlated with tumor progression and prognosis in Chinese breast cancer patients. IUBMB Life 59: 450–457. DOI 10.1080/15216540701509955. [Google Scholar] [CrossRef]
Zhang M, Jin X, Zhang Z, Li B, Yang G (2018). Vildagliptin protects endothelial cells against high glucose-induced damage. Biomedicine & Pharmacotherapy 108: 1790–1796. DOI 10.1016/j.biopha.2018.09.148. [Google Scholar] [CrossRef]
Zheng H, You H, Zhou XZ, Murray SA, Uchida T et al. (2002). The prolyl isomerase Pin1 is a regulator of p53 in genotoxic response. Nature 419: 849–853. DOI 10.1038/nature01116. [Google Scholar] [CrossRef]
Zheng W, Zhou J, Song S, Kong W, Xia W et al. (2018). Dipeptidyl-peptidase 4 inhibitor sitagliptin ameliorates hepatic insulin resistance by modulating inflammation and autophagy in ob/ob mice. International Journal of Endocrinology 2018: 8309723. DOI 10.1155/2018/8309723. [Google Scholar] [CrossRef]
Zong WX, Rabinowitz JD, White E (2016). Mitochondria and cancer. Molecular Cell 61: 667–676. DOI 10.1016/j.molcel.2016.02.011. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2022 The Author(s). Published by Tech Science Press.
Copyright © 2022 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools