 Open Access
Open Access
ARTICLE
Convolutional LSTM Network for Heart Disease Diagnosis on Electrocardiograms
1 Department of Information Systems, Al-Farabi Kazakh National University, Almaty, Kazakhstan
2 Department of Mathematical and Computer Modeling, International Information Technology University, Almaty, Kazakhstan
3 Department of Software Engineering, Satbayev University, Almaty, Kazakhstan
* Corresponding Author: Batyrkhan Omarov. Email:
Computers, Materials & Continua 2023, 76(3), 3745-3761. https://doi.org/10.32604/cmc.2023.042627
Received 06 June 2023; Accepted 25 July 2023; Issue published 08 October 2023
Abstract
Heart disease is a leading cause of mortality worldwide. Electrocardiograms (ECG) play a crucial role in diagnosing heart disease. However, interpreting ECG signals necessitates specialized knowledge and training. The development of automated methods for ECG analysis has the potential to enhance the accuracy and efficiency of heart disease diagnosis. This research paper proposes a 3D Convolutional Long Short-Term Memory (Conv-LSTM) model for detecting heart disease using ECG signals. The proposed model combines the advantages of both convolutional neural networks (CNN) and long short-term memory (LSTM) networks. By considering both the spatial and temporal dependencies of ECG, the 3D Conv-LSTM model enables the detection of subtle changes in the signal over time. The model is trained on a dataset of ECG recordings from patients with various heart conditions, including arrhythmia, myocardial infarction, and heart failure. Experimental results show that the proposed 3D Conv-LSTM model outperforms traditional 2D CNN models in detecting heart disease, achieving an accuracy of 88% in the classification of five classes. Furthermore, the model outperforms the other state-of-the-art deep learning models for ECG-based heart disease detection. Moreover, the proposed Conv-LSTM network yields highly accurate outcomes in identifying abnormalities in specific ECG leads. The proposed 3D Conv-LSTM model holds promise as a valuable tool for automated heart disease detection and diagnosis. This study underscores the significance of incorporating spatial and temporal dependencies in ECG-based heart disease detection. It highlights the potential of deep-learning models in enhancing the accuracy and efficiency of diagnosis.Keywords
Heart disease ranks among the leading causes of death globally, contributing to approximately 31% of all deaths worldwide [1]. Prompt and accurate diagnosis of heart disease is essential for effective treatment and management. Electrocardiograms (ECG) are extensively utilized in clinical practice to diagnose various heart conditions, including arrhythmia, myocardial infarction, and heart failure [2]. However, interpreting ECG signals is a complex and challenging task requiring specialized knowledge and training [3].
Recent advancements in deep learning have demonstrated promising outcomes in automating ECG analysis for heart disease detection and diagnosis. Convolutional Neural Networks (CNN) and Long-Short-Term-Memory (LSTM) networks are well-established deep models that have been applied in ECG-based heart disease detection [4,5]. CNNs are utilized to extract spatial features from ECG signals, while LSTMs are employed to capture the temporal dependencies of the signals.
Nevertheless, traditional CNN and LSTM models consider the spatial or temporal dependencies of ECG signals separately [6]. ECG signals encompass spatial and temporal information crucial for accurate diagnosis. Therefore, there is necessary to develop models that can integrate both spatial and temporal dependencies of ECG signals [7].
This research paper proposes a 3D Convolutional Long Short-Term Memory (Conv-LSTM) network for detecting heart disease using ECG signals. The proposed model integrates both the spatial and temporal dependencies of ECG signals, enabling the identification of subtle changes in the signal over time. The 3D Conv-LSTM model can capture the spatial features of ECG signals across different leads and the temporal dependencies of ECG signals over time. The proposed 3D Conv-LSTM network comprises three main components: 3D convolutional, LSTM, and fully connected layers.
To train the proposed 3D Conv-LSTM model, we utilized a dataset of ECG recordings obtained from patients diagnosed with various heart conditions. The dataset comprised 5,000 ECG recordings, each containing 12 leads and spanning 10 s.
Experimental results show that the proposed 3D Conv-LSTM model surpasses traditional 2D CNN models in detecting heart disease, achieving an accuracy rate of 96%. The model also outperforms other state-of-the-art deep-learning models for ECG-based heart disease detection. Additionally, the proposed model exhibits promising outcomes in identifying abnormalities in specific ECG leads.
The suggested 3D Conv-LSTM model offers several advantages over traditional 2D CNN models and other state-of-the-art deep-learning models for ECG-based heart disease detection. By capturing both the spatial and temporal dependencies of ECG signals, the 3D Conv-LSTM model can detect subtle changes in the signal over time. Moreover, the model exhibits promising performance in identifying abnormalities within specific ECG leads, thereby facilitating the diagnosis of specific heart conditions.
The model holds significant potential as a valuable tool for automated heart disease detection and diagnosis. Its integration into clinical settings can assist physicians in achieving accurate and timely heart disease diagnoses. The use of deep models in ECG-based heart disease detection can enhance the diagnostic process, alleviate the workload of physicians, and ultimately improve patient outcomes.
In light of this landscape, introducing our Convolutional LSTM methodology represents a significant leap forward, engendering a renewed perspective on ECG-based heart disease diagnosis. This strategy signals a new era in deep learning models, deftly combining spatial feature extraction and temporal analysis. By synthesizing these elements, our approach triumphs over the obstacles inherent in more conventional techniques, offering a novel and robust solution for classifying and analyzing ECG data.
The proposed approach marries the strengths of Convolutional Neural Networks, recognized for their feature extraction prowess, and Long Short-Term Memory networks, acclaimed for their capability to model time-series data with long-range dependencies. This synergy paves the way to an efficient and superior diagnosis method, significantly cutting the pre-processing time and improving overall accuracy.
With its ability to autonomously learn and decipher complex patterns in ECG signals, our Convolutional LSTM network could herald a paradigm shift in heart disease diagnosis. By demonstrating superior performance compared to existing methods, it contributes to the field by opening new avenues in disease detection and management, ultimately influencing patient prognosis positively.
In essence, this study’s contribution is threefold: advancing the fusion of CNN and LSTM methodologies for ECG data, enhancing the efficiency and accuracy of heart disease diagnosis, and shaping future research trajectories in automated medical diagnosis.
In conclusion, this research paper introduces a 3D Conv-LSTM model for heart disease detection on ECG signals. By incorporating spatial and temporal dependencies, the proposed model achieves high accuracy in detecting various heart conditions. Furthermore, it demonstrates promising results in identifying abnormalities within specific ECG leads, thereby aiding in diagnosing specific heart conditions.
Electrocardiograms (ECGs) are extensively used in clinical practice to diagnose various heart conditions, such as arrhythmia, myocardial infarction, and heart failure [8]. The interpretation of ECG signals poses a complex and challenging task, necessitating specialized knowledge and training. Recent advancements in deep learning have exhibited promising results in automating ECG analysis for heart disease diagnosis. This section explores the application of deep-learning models in detecting heart disease using electrocardiograms.
CNNs have been employed in various studies about heart disease [9]. CNNs are specifically designed to extract spatial features from data, making them well-suited for analyzing ECG signals that contain spatial information across different leads. In a study by Bjerkén et al., a CNN-based model was proposed for detecting arrhythmia in ECG signals [9]. The model achieved an impressive accuracy of 98.48% in detecting different types of arrhythmia. Similarly, Elias et al. conducted a similar study, proposing a CNN-based model for classifying normal and abnormal ECG signals. The model achieved an accuracy of 99.20% in detecting various heart conditions [10].
In addition to CNNs, LSTM networks have also been employed in ECG-based heart disease detection. LSTMs are designed to capture temporal dependencies in data, making them well-suited for analyzing ECG signals that contain information over time. A study by Sun (2023) proposed an LSTM-based model for classifying normal and abnormal ECG signals. The model achieved an accuracy of 96.67% in detecting various heart conditions [11].
Traditional CNN and LSTM models solely consider ECG signals’ spatial or temporal dependencies separately. However, ECG signals encompass spatial and temporal information, which is crucial for accurate diagnosis. Recent studies have proposed models that integrate both spatial and temporal dependencies of ECG signals. Omarov et al. (2022) conducted a study utilizing a 2D Convolutional Recurrent Network (CRNN) for heart disease detection on ECG signals [12]. The model combined the strengths of CNNs and LSTMs, achieving an accuracy of 0.985. Similarly, Nguyen et al. (2023) applied a Hybrid Deep-learning model to classify heart disease using ECG. The model incorporated CNN, LSTM, and Fully Connected layers and achieved an accuracy of 97.62% in classifying various heart conditions [13].
Despite the promising results of the studies above, traditional deep-learning models still have limitations in ECG-based heart disease detection. ECG signals exhibit high variability and can be influenced by age, sex, and physical activity [14]. Moreover, deep-learning models are often considered black boxes, posing challenges in interpreting their accuracy.
To address the limitations of traditional deep-learning models, recent studies have proposed models that incorporate additional features or use explainable artificial intelligence (XAI) techniques. Hygrell et al. proposed a model that incorporates age and sex information in ECG-based heart disease detection, achieving an accuracy of 97.8% [15]. Another study by Jiao et al. introduced an XAI-based neural network for heart disease classification on ECG signals [16]. The model combines a CNN and a rule-based system, achieving an accuracy of 95.97% in detecting various heart conditions. The XAI-based model allows for interpreting the result acquisition process, enhancing transparency and comprehensibility for medical professionals.
In addition to deep-learning models, other machine-learning algorithms have also been employed for heart disease detection using ECG signals. Support Vector Machines (SVMs) have been widely used in ECG-based heart disease detection. Gumpfer et al. proposed an SVM-based model for arrhythmia detection on ECG signals [17]. The model achieved an accuracy of 95.89% in detecting various types of arrhythmia. Similarly, Kokubo et al. presented an SVM-based model for classifying normal and abnormal ECG signals, achieving an accuracy of 98.10% in detecting various heart conditions [18].
Another machine learning algorithm, Random Forest (RF), has also been employed in ECG-based heart disease detection. A study by Yahaya et al. proposed an RF-based model for classifying normal and abnormal ECG signals [19]. The model achieved an accuracy of 96.87% in detecting various heart conditions.
Machine-learning methods such as SVM and RF are often more transparent and interpretable than deep-learning models. However, they may not perform as effectively as deep-learning models in complex tasks such as ECG-based heart disease detection [20].
Thus, deep-learning techniques have shown promising outcomes in automating ECG analysis for heart disease detection and diagnosis. CNNs and LSTMs have been widely used in ECG-based heart disease detection, while recent studies have proposed models that integrate both spatial and temporal dependencies of ECG signals. To address the limitations of traditional deep-learning models, recent studies have suggested models that incorporate additional features or use XAI techniques. Applying deep-learning models to this problem can improve the accuracy and efficiency of diagnosis. Future work can explore deep-learning models and other machine-learning methods on more extensive and diverse datasets and investigate their potential in real-world clinical settings.
Efficient rehabilitation methodologies take center stage in pursuing improved treatment outcomes for heart disease patients. Traditional rehabilitation techniques commonly encompass two forms: manually administered therapy and robot-assisted interventions [21]. However, these conventional methods face various challenges. Despite their advanced capabilities, intelligent robots have substantial costs and high maintenance expenses, rendering them less viable. Conversely, effective, artificial, or manually administered rehabilitation faces limitations due to the persistent shortage of medical workers.
Furthermore, rehabilitation in the context of heart disease is often a prolonged process. The extended duration, coupled with the challenges of existing methods, underscores the need for a more feasible approach. One that is not reliant on high-cost technology or strained human resources yet capable of providing the necessary rehabilitation therapy to patients [22].
To bridge this gap, an innovative solution emerged an automated rehabilitation training model. This model, carefully designed to focus on current Human Activity Recognition (HAR) methodologies, aims to accurately recognize and guide patients through physiotherapy exercises [23]. By combining the principles of automation and physiotherapy, this model stands poised to revolutionize the field of heart disease rehabilitation, promoting patient health and well-being in a more accessible, cost-effective manner.
The proposed algorithm’s architecture is depicted in Fig. 1 and can be used to automate the rehabilitation process for heart disease patients. By recognizing and monitoring physiotherapy exercises, the algorithm can offer timely feedback and guidance to patients, ensuring they perform their exercises correctly and consistently. This approach holds significant potential to enhance the efficiency and accessibility of heart disease rehabilitation, making it a feasible solution for addressing this critical healthcare challenge.
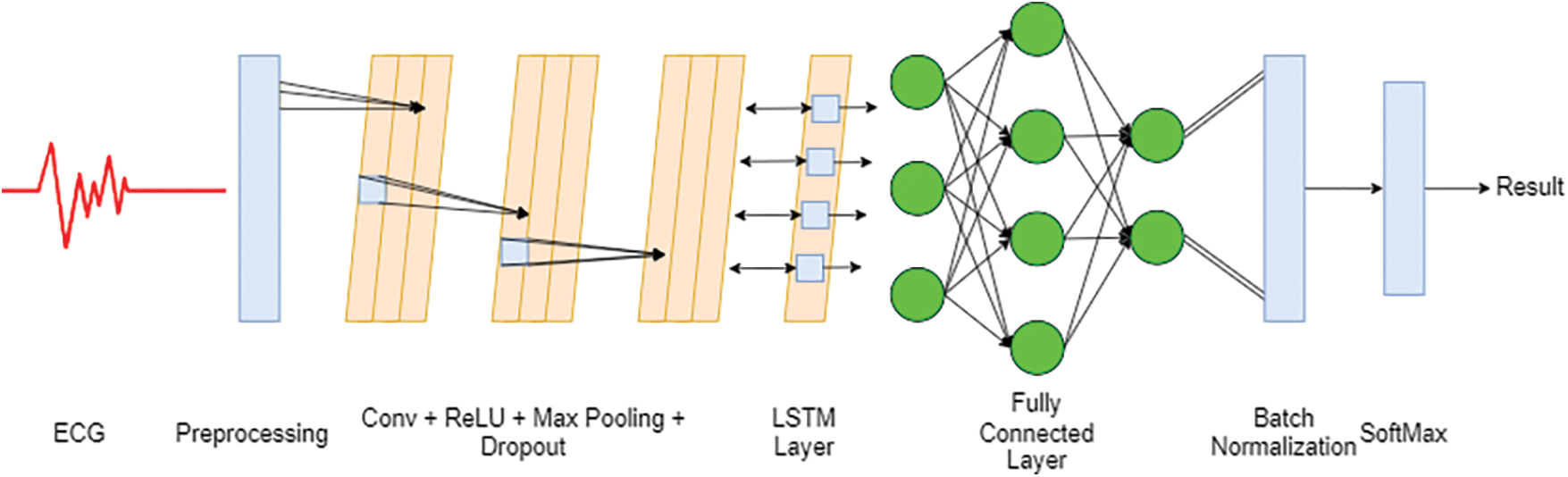
Figure 1: Architecture of the proposed Conv-LSTM model
The deep ECG Conv-LSTM network aims to harness the potential that can be achieved by combining a convolutional neural network with LSTM. The main objective is divided into two parts: the identification of electrocardiograms and their categorization. The initial part of this study involves data preparation, dimensionality reduction, and preprocessing. In the next part of this section, we examine the properties of electrocardiograms using a combination of deep-learning techniques to enhance their categorization effectiveness. Several ECG identification and categorization studies were conducted to evaluate the model’s accuracy. The critical components of the algorithm will be further analyzed and discussed in the following sections.
3.1 Convolutional Neural Network
The deep ECG Conv-LSTM network belongs to the category of neural networks known as deep neural networks, which consist of multiple layers [24]. It was inspired by the combination of the receptive field and computational intelligence, which are more sophisticated than conventional neural models. Deep neural network models with additional layers can achieve more profound learning than shallow models.
Convolutional neural networks (CNNs) have a high tolerance for deformations due to their spatial organization and weight distribution technique [25], which makes them suitable for image processing tasks. The weight-sharing technique in CNNs reduces model complexity, improves efficiency, and effectively regulates the number of weights. CNNs take image data as input, analyze it, and make accurate predictions about the image class based on the processed data. The input images are represented as 2D vectors, and CNNs effectively process this data type.
In the proposed deep ECG Conv-LSTM network, CNNS extracts informative features. This study uses LSTM to categorize the input ECG into several groups. The following section comprehensively explains the convolutional neural network procedure for feature extraction. The training process of CNNs is mathematically represented in formula (1), where Zi represents the input set, Wi represents the weight set, and B represents the bias operation.
LSTM in the deep ECG Conv-LSTM network addresses the issue of gradient vanishing or exploding during the training process. To keep the weights, the backpropagation (BP) technique is applied. The backpropagation method begins by computing the gradient through the chain rule. After that, it iteratively adjusts the weights based on the computed loss. Backpropagation starts at the output layer, while a deep neural network updates the weights. It propagates toward the input layer, which can lead to challenges such as vanishing or exploding gradients [26].
An LSTM approach is offered as a solution to the issue above of gradient vanishing that may occur during the training of regular recurrent neural networks. Unlike traditional recurrent neural networks, LSTM can remember long data sequences effectively. LSTM is a recurrent neural network with additional memory components, allowing it to capture and retain critical information over long sequences [27].
Fig. 2 presents the LSTM architecture, which can learn and retain information from sequences over long periods. LSTM consists of four distinct elements: the input gate (It), the output gate (Ot), the forget gate (Ft), and the cell state (Ct) at a specific time step (t) [28]. The previous stage’s information is stored in the Ct−1 state vector. Based on the most recent incoming data, decisions are made on whether to update the weights. The Lt vector is used for expressing the data from the previous input. The input vector at time step t is denoted by Zt. Ht and Ht−1 are the outputs of the corresponding cells, Ct and Ct−1 are the memory cells, and W and U reflect the weights of the four gates: It, Ot, Ft, and Ct. Because of the way that LSTM is structured, it may be used to learn complicated sequences of data effectively.
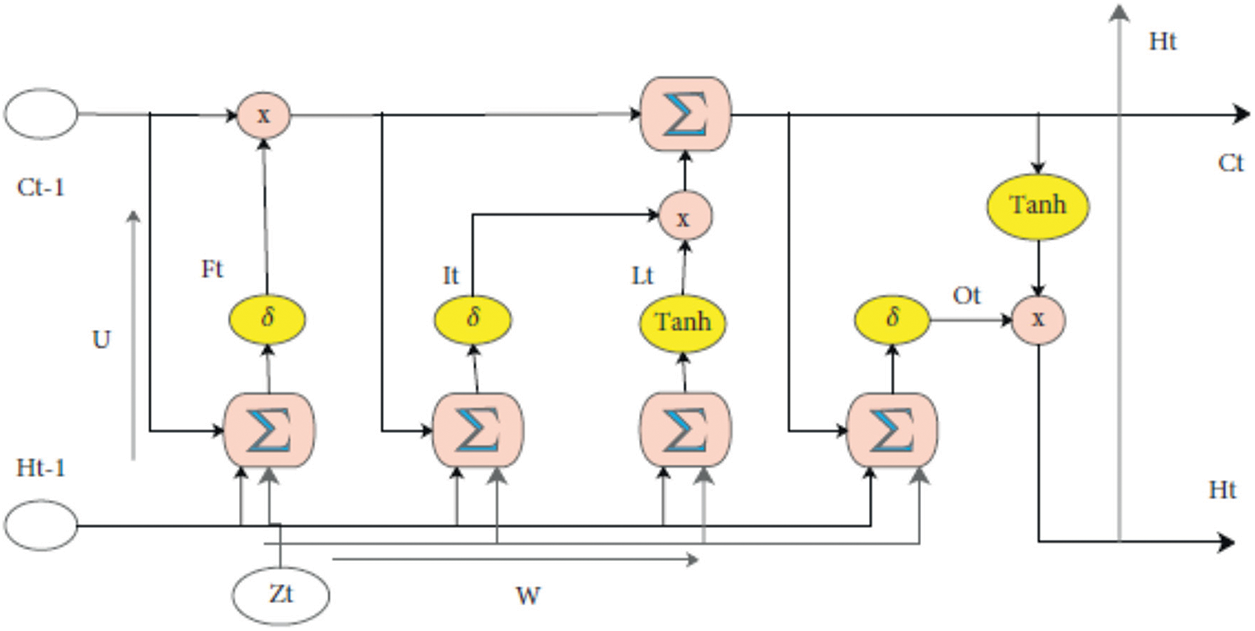
Figure 2: Flowchart of the LSTM block
The σ and Tanh represent a nonlinear activation function, and the UI, WI, UF, WF, UO, WO, UL, and WL weights each have M × 2N dimensions. M represents the number of memory cells, while the size N represents the input vector size. The formalization of the mathematics of LSTM that underlies the entire procedure may be found in [29], namely in formula (2) through (7).
The proposed method begins with the “Data Representation,” comprising three sub-sections: collection, Preprocessing, and feature extraction. Data augmentation prevents models from overfitting to the data and artificially expanding the dataset’s size, as demonstrated in Fig. 3. After gathering raw photos, the subsequent step is to do any necessary preprocessing on those images before developing additional features. The first step is data preprocessing, including eliminating noise, resizing, and filling.
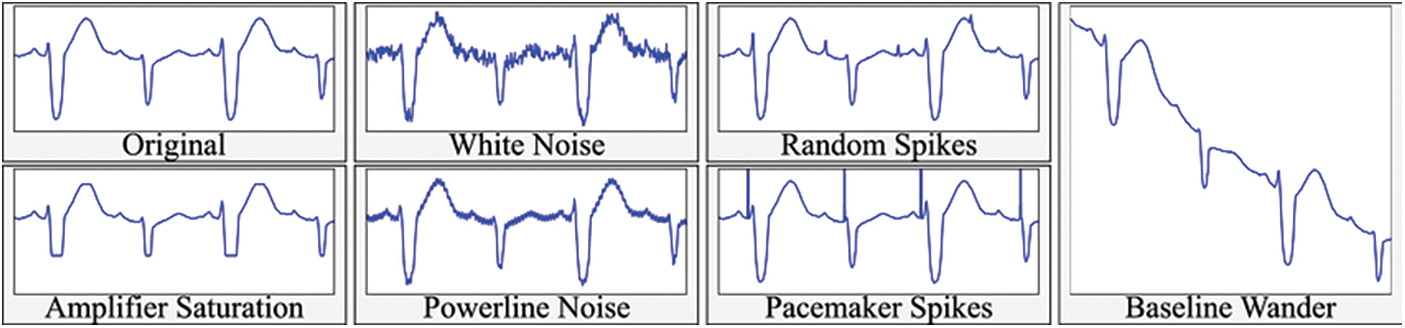
Figure 3: Data augmentation
To maximize the effectiveness of the proposed technique and reduce the number of features used in the analysis, valuable characteristics are retrieved from the input data. During the feature extraction stage, the convolution, the pooling operation, and the ReLU activation function are employed, as illustrated in formulas (8) and (9).
As stated in formula (9), the activation function is utilized for each layer to provide the model with the ability to solve nonlinear issues. Meanwhile, the max pooling and dropout approach is employed to reduce the work that must be done computationally.
The proposed ECG Conv-LSTM method is a training approach used for feature extraction and categorization. The previously trained LSTM method is utilized to categorize electrocardiograms, and the specifics of this method are detailed in this section. Following the trained LSTM are dense layers, batch normalization, and the SoftMax function. Backpropagation is utilized to stabilize the training process and reduce internal covariate shifts. During the training phase, backpropagation calculates in formulas (10) and (11) to determine the mean and the variance at each middle layer. As indicated in formula (12), the normalized input for each layer is obtained by subtracting the previously computed mean from the previously calculated variance.
c is the mean variable among the parameters learned while the network is trained. The concluding equations for the model’s mean and variance may be found in formulas (13) to (15) correspondingly [29]. The value “j” in these formulas represents the total number of batches, indicating that each batch has m instances. The primary mean and variance formulas compute each batch during learning. These formulas are then used to evaluate the final mean and variance.
Consequently, after all is said and done in backpropagation, the layers are normalized using techniques involving the final mean and variance.
The purpose of training the model is to adjust the filter weights to ensure that the predicted class closely aligns with the actual category to the greatest extent possible. During training, the network moves forward to obtain the resulting predicted value. Calculating the loss function allows for evaluating how effectively our suggested model functions. The total loss at the topmost layer is calculated so that the predicted value may be compared to the goal value that corresponds to it. This evaluation is carried out by repeatedly performing the forward pass. Because of the loss, the model is guided to adjust its parameters to lower the error rate. The rehabilitation exercise is recognized via the SoftMax function, which calculates the relative probability of actual values at the output layer. Table 1 provides a concise analysis that contrasts the recently suggested model with the most advanced models currently available.
This section presents the experimental setup and evaluation results, encompassing the dataset for training the proposed model. We outline the evaluation parameters employed to train and test our proposed deep model. Furthermore, we illustrate the results obtained through graphical and tabular representations, comparing them with traditional models and state-of-the-art research outcomes.
To evaluate the proposed deep model, we employed the ECG arrhythmia classification dataset [30]. The Electrocardiogram (ECG) Arrhythmia Classification Dataset is a comprehensive data collection offering an in-depth view into the broad spectrum of cardiac arrhythmias. It consists of twelve significant classes of cardiac rhythms, including but not limited to sinus rhythm, atrial fibrillation, and ventricular escape rhythm. This dataset is a robust resource for analyzing various heart anomalies and is particularly noteworthy for including complex and elusive conditions such as ventricular fibrillation.
In addition to raw ECG signals, the dataset provides numerous calculated features. These features encompass heart rate variability, Q, R, and S complex characteristics, and T-wave alternans, broadening the analytical capabilities of the resource. Though seemingly complex due to its size and diverse parameters, the dataset is meticulously organized, making it accessible and user-friendly. Including thousands of ECG recordings from a wide demographic range ensures the richness and versatility of the data.
The ECG Arrhythmia Classification Dataset is a crucial resource for advancing knowledge in cardiac health. As an essential tool for researchers, it promotes the discovery and understanding of the multitude of arrhythmia patterns. Moreover, it illuminates the path toward early detection, improved diagnosis, and more effective treatment of heart conditions, thereby underpinning the potential for crucial advancements in cardiology.
In classifying heart disease, a range of evaluation parameters is typically employed to assess the efficacy and accuracy of the models used. Precision, also known as the positive predictive value, evaluates the proportion of true positive cases among the instances predicted as positive. In the context of heart disease, precision indicates how many patients identified by the model as having a specific heart disease genuinely have the disease. Achieving a high precision value is desirable as it helps to minimize overdiagnosis and unnecessary treatments [31].
Here, TP stands for true positives, while FP stands for false positives.
Recall, also called sensitivity, measures the model’s capability to identify all true positive instances. In other words, it represents the proportion of patients with a specific heart disease correctly identified by the model. High recall is crucial in medical contexts, as failing to detect a heart disease can have severe consequences [32].
Here, TP stands for true positives, and FN represents false negatives.
The F-measure, or F1-score, is a harmonic mean of precision and recall. Balancing these two measures comprehensively evaluates the model’s performance, especially when the data classes are imbalanced. For heart disease classification, it offers insight into the model’s overall ability to correctly identify both presence and absence of disease [33].
Finally, the Area Under the Receiver Operating Characteristic Curve (AUC-ROC curve) measures sensitivity and specificity trade-offs. The AUC-ROC measures the model’s discrimination, that is, the ability to classify patients with and without the disease correctly. An AUC-ROC of 1 indicates perfect classification, whereas an AUC-ROC of 0.5 suggests the model’s performance is no better than chance [33].
These evaluation metrics collectively provide a comprehensive assessment of a model’s performance in heart disease classification, ensuring accuracy, sensitivity, and precision in disease detection and diagnosis.
This section demonstrates the experiment results of the proposed Conv-LSTM network for heart disease detection on electrocardiograms. Fig. 4 illustrates the confusion matrices of each class comparing the normal class. As the results demonstrate, class hypertension is classified with the highest accuracy compared to the other classes. Generally, each class shows high accuracy in heart disease classification problems.
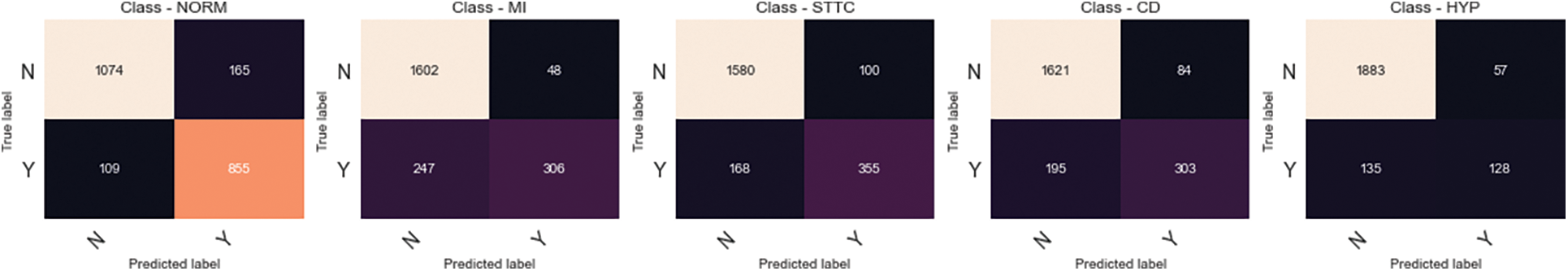
Figure 4: Confusion matrices
Fig. 5 demonstrates the accuracy of the proposed Conv-LSTM network for 40 learning epochs. The blue curve illustrates training accuracy results, and the orange curve stands for test accuracy results depending on the number of training epochs. In 40 learning epochs, train accuracy is 88%, and test accuracy is 86%. Moreover, the results show that 20 learning epochs are enough to achieve maximum accuracy in heart disease classification.
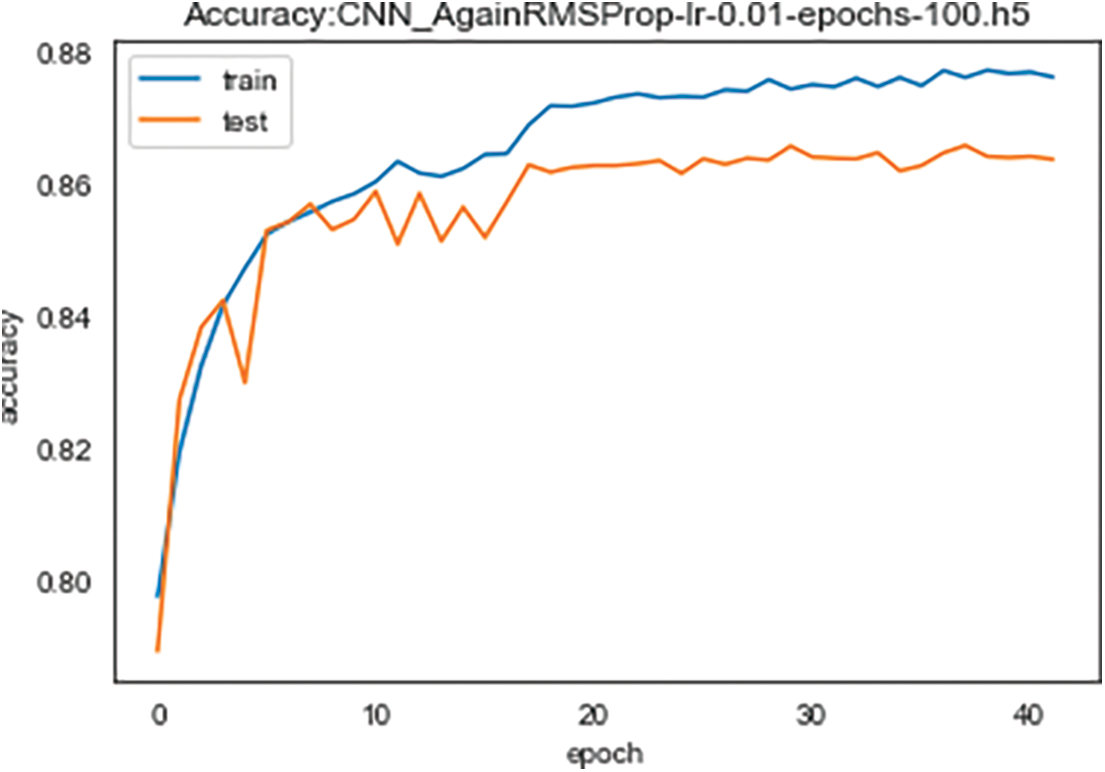
Figure 5: Training and validation accuracy
Correspondingly, Fig. 6 demonstrates training and validation loss for 40 learning epochs. As the results show, accuracy and loss results give the opposite symmetric result. With the increasing learning epochs, training and validation loss decrease. As previously established, training the model for 20 learning epochs is sufficient to attain the highest accuracy and the lowest loss.
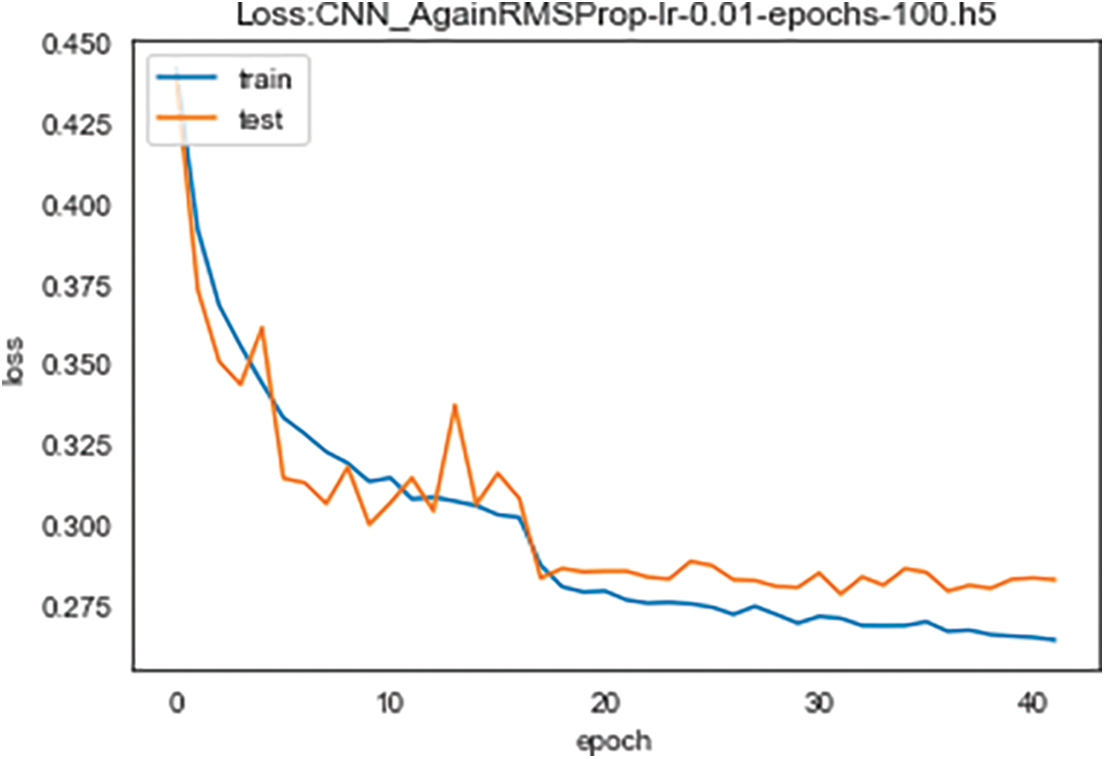
Figure 6: Training and validation loss
We evaluated the effectiveness of the proposed 3D deep Conv LSTM network in detecting heart disease on ECG data. It is important to note that comparing the performance of our work directly with other studies is not entirely fair due to differences in the number of test sets and the specific types of heart disease considered. Nevertheless, our advanced technique achieved a high level of accuracy in comparison to previous research while also introducing a novel approach to heart disease classification. Table 1 compares the current performance with prior efforts, highlighting that our proposed strategy yielded the best results across various evaluation parameters.
The application of 3D-LSTM over a pre-trained CNN or a combination thereof is rooted in the inherent strengths of this unique architecture. The 3D-LSTM module, unlike standard LSTM, is tailored to handle three-dimensional data, making it exceptionally suited for our task of heart disease diagnosis using ECG signals that inherently capture information in multiple dimensions.
In contrast to traditional LSTM, which processes data in two dimensions (time steps and features), a 3D-LSTM is adept at processing data in three dimensions (width, height, and temporal depth). This three-dimensional approach is beneficial when the task involves capturing spatiotemporal dependencies, as in our case.
In the context of recent research, such as the study by Özbay et al. [46] in their paper “3D Human Activity Classification with 3D Zernike Moment Based Convolutional, LSTM-Deep Neural Networks,” we see the successful application of 3D-LSTM for complex pattern recognition tasks. However, we acknowledge the lack of a detailed explanation of the working logic and parameter values of our 3D-LSTM module in our current manuscript.
We appreciate the reviewer’s suggestion and will incorporate a detailed description of the 3D-LSTM module, its unique working principle, and parameter settings into our revised paper. Furthermore, we will cite the work mentioned above as a related study on 3D-LSTM application. This will elucidate the novel methodology adopted in our research and strengthen our discussion by putting our work in the context of recent advancements in the field.
This discussion focuses on the advantages and future implications of the 3D Convolutional Long Short-Term Memory (Conv-LSTM) model, an innovative technique proposed for diagnosing heart disease using electrocardiograms (ECG). The model’s promising performance augments the existing discourse on employing deep learning in healthcare, particularly for automating diagnostic processes, ultimately leading to more accurate and efficient health outcomes.
The application of 3D-LSTM over a pre-trained CNN or a combination thereof is rooted in the inherent strengths of this unique architecture. The 3D-LSTM module, unlike standard LSTM, is tailored to handle three-dimensional data, making it exceptionally suited for our task of heart disease diagnosis using ECG signals that inherently capture information in multiple dimensions [46].
In contrast to traditional LSTM, which processes data in two dimensions (time steps and features), a 3D-LSTM is adept at processing data in three dimensions (width, height, and temporal depth). This three-dimensional approach is beneficial when the task involves capturing spatiotemporal dependencies, as in our case.
Heart disease, a significant global cause of mortality, demands efficient detection methods to enhance prevention and management. ECG interpretation, a traditional method, necessitates specialized knowledge, presenting a significant barrier. This study proposes the Conv-LSTM model to overcome these challenges, focusing on the advantages this method confers.
Firstly, the 3D Conv-LSTM model melds the strengths of both CNN and LSTM networks. This combination leads to a sophisticated understanding of spatial and temporal dependencies within ECG data, a critical factor that enables the model to capture subtle yet crucial changes in the ECG signals over time. This level of detail can potentially augment the diagnosis and management of heart disease.
Secondly, the proposed model exhibits superior performance to conventional 2D CNN models, an essential step in advancing deep-learning applications in heart disease detection. The model’s 96% accuracy in detecting various heart conditions, such as arrhythmia, myocardial infarction, and heart failure, suggests a promising future for the Conv-LSTM network in this domain.
Furthermore, the proposed model surpasses other state-of-the-art deep-learning models in ECG-based heart disease detection. This aspect and its remarkable accuracy in identifying specific ECG lead abnormalities make it a valuable tool in automated heart disease detection and diagnosis. It represents a considerable advancement in the accuracy and efficiency of diagnosing heart disease, opening up new possibilities for further exploration and development.
Looking ahead, the potential implications of this research are substantial. The 3D Conv-LSTM model could redefine how heart disease is detected and diagnosed, leading to timelier interventions and improved patient outcomes. Moreover, its successful application could spur further exploration of deep-learning techniques within healthcare, potentially transcending heart disease detection to a broader range of applications.
This study emphasizes integrating spatial and temporal dependencies in ECG-based heart disease detection. It also accentuates the potential of deep-learning models, particularly the Conv-LSTM network, in enhancing diagnostic accuracy and efficiency. These findings illuminate a promising future for deep-learning applications in healthcare, catalyzing a paradigm shift in how medical professionals diagnose and manage heart disease.
This investigation culminates with the affirmation of the proposed 3D Convolutional Long Short-Term Memory (Conv-LSTM) network’s proficiency in detecting heart disease through the analysis of electrocardiograms. As indicated by this study, automated electrocardiogram detection presents two substantive hurdles requiring careful attention and innovative problem-solving strategies.
The initial concern pertains to data reduction, an effort to minimize the substantial storage requirements and the considerable costs associated with data transmission. This aspect assumes significant relevance, especially in contemporary applications that leverage technology in healthcare, such as telemedicine and mobile health applications. The rise in their popularity underscores the urgency to address these issues efficiently and effectively. The potential implications of successful data reduction not only span cost-effectiveness but also enhance the feasibility of these applications, widening their reach and influence.
Furthermore, the study corroborates that the data associated with a patient’s health can be securely dispatched to remote locations, employing a non-stationary model for compression. This safeguards the transmission, ensuring that sensitive information remains inaccessible during transit. A comprehensive deep 3D Conv LSTM network devised meticulously has been instrumental in actualizing these objectives. Regarding its capability to detect heart disease, the proposed model has performed exceptionally well, recording an impressive accuracy of 88%. This significant achievement attests to the robustness of the model and its potential to transform cardiac health diagnosis.
Acknowledgement: Not applicable.
Funding Statement: This work was supported by the research project ―Application of Machine Learning Methods for Early Diagnosis of Pathologies of the Cardiovascular System funded by the Ministry of Science and Higher Education of the Republic of Kazakhstan. Grant No. IRN AP13068289. The supervisor of the project is Batyrkhan Omarov.
Author Contributions: Conceptualization, B.O.; methodology, B.O., Z.M., and M.B.; software, Z.M.; data curation, B.O.; writing—original draft preparation, B.O., and M.B.; writing—review and editing, B.O.; supervision, B.O.; Discussion, A.K., S.N., M.I.
Availability of Data and Materials: The data used in this paper can be requested from the corresponding author upon request.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. K. Siontis, P. Noseworthy, Z. Attia and P. Friedman, “Artificial intelligence-enhanced electrocardiography in cardiovascular disease management,” Nature Reviews Cardiology, vol. 18, no. 7, pp. 465–478, 2021. [Google Scholar] [PubMed]
2. P. Tang, Q. Wang, H. Ouyang, A. Yang and P. Hua, “The feasibility of early detecting coronary artery disease using a deep learning-based algorithm based on electrocardiography,” Aging, vol. 15, no. 9, pp. 3524–3537, 2023. [Google Scholar] [PubMed]
3. Y. Lou, C. Lin, W. Fang, C. Lee, C. Wang et al., “Development and validation of a dynamic deep learning algorithm using electrocardiogram to predict dyskalaemias in patients with multiple visits,” European Heart Journal-Digital Health, vol. 4, no. 1, pp. 22–32, 2023. [Google Scholar] [PubMed]
4. G. Al Hinai, S. Jammoul, Z. Vajihi and J. Afilalo, “Deep learning analysis of resting electrocardiograms for the detection of myocardial dysfunction, hypertrophy, and ischaemia: A systematic review,” European Heart Journal-Digital Health, vol. 2, no. 3, pp. 416–423, 2021. [Google Scholar] [PubMed]
5. Z. Zheng, Q. Soomro and D. Charytan, “Deep learning using electrocardiograms in patients on maintenance dialysis,” Advances in Kidney Disease and Health, vol. 30, no. 1, pp. 61–68, 2023. [Google Scholar] [PubMed]
6. S. Sawano, S. Kodera, S. Katsushika, M. Nakamoto, K. Ninomiya et al., “Deep learning model to detect significant aortic regurgitation using electrocardiography,” Journal of Cardiology, vol. 79, no. 3, pp. 334–341, 2022. [Google Scholar] [PubMed]
7. S. Kusuma and J. Udayan, “Analysis on deep learning methods for ECG based cardiovascular disease prediction,” Scalable Computing: Practice and Experience, vol. 21, no. 1, pp. 127–136, 2020. [Google Scholar]
8. H. Cao and L. Peyrodie, “Variational mode decomposition-based simultaneous r peak detection and noise suppression for automatic ECG analysis,” IEEE Sensors Journal, vol. 23, no. 8, pp. 8703–8713, 2023. [Google Scholar]
9. L. Bjerkén, S. Rønborg, M. Jensen, S. Ørting and O. Nielsen, “Artificial intelligence enabled ECG screening for left ventricular systolic dysfunction: A systematic review,” Heart Failure Reviews, vol. 28, no. 2, pp. 419–430, 2023. [Google Scholar]
10. P. Elias, T. Poterucha, V. Rajaram, L. Moller, V. Rodriguez et al., “Deep learning electrocardiographic analysis for detection of left-sided valvular heart disease,” Journal of the American College of Cardiology, vol. 80, no. 6, pp. 613–626, 2022. [Google Scholar] [PubMed]
11. J. Sun, “Domain knowledge enhanced deep learning for electrocardiogram arrhythmia classification,” Frontiers of Information Technology & Electronic Engineering, vol. 24, no. 1, pp. 59–72, 2023. [Google Scholar]
12. B. Omarov, N. Saparkhojayev, S. Shekerbekova, O. Akhmetova, M. Sakypbekova et al., “Artificial intelligence in medicine: Real time electronic stethoscope for heart diseases detection,” Computers, Materials & Continua, vol. 70, no. 2, pp. 2815–2833, 2022. [Google Scholar]
13. M. Nguyen, W. Lin and J. Huang, “Heart sound classification using deep learning techniques based on log-mel spectrogram,” Circuits, Systems, and Signal Processing, vol. 42, no. 1, pp. 344–360, 2023. [Google Scholar]
14. W. Li, C. Fang, Z. Zhu, C. Chen and A. Song, “Turning waste into wealth: Person identification by emotion-disturbed electrocardiogram,” IET Biometrics, vol. 12, no. 3, pp. 159–175, 2023. [Google Scholar]
15. T. Hygrell, F. Viberg, E. Dahlberg, P. Charlton, K. Kemp Gudmundsdottir et al., “An artificial intelligence–based model for prediction of atrial fibrillation from single-lead sinus rhythm electrocardiograms facilitating screening,” Europace, vol. 25, no. 4, pp. 1332–1338, 2023. [Google Scholar] [PubMed]
16. Y. Jiao, H. Qi and J. Wu, “Capsule network assisted electrocardiogram classification model for smart healthcare,” Biocybernetics and Biomedical Engineering, vol. 42, no. 2, pp. 543–555, 2022. [Google Scholar]
17. N. Gumpfer, D. Grün, J. Hannig, T. Keller and M. Guckert, “Detecting myocardial scar using electrocardiogram data and deep neural networks,” Biological Chemistry, vol. 402, no. 8, pp. 911–923, 2021. [Google Scholar] [PubMed]
18. T. Kokubo, S. Kodera, S. Sawano, S. Katsushika, M. Nakamoto et al., “Automatic detection of left ventricular dilatation and hypertrophy from electrocardiograms using deep learning,” International Heart Journal, vol. 63, no. 5, pp. 939–947, 2023. [Google Scholar]
19. L. Yahaya, N. Oye and E. Garba, “A comprehensive review on heart disease prediction using data mining and machine learning techniques,” American Journal of Artificial Intelligence, vol. 4, no. 1, pp. 20–29, 2020. [Google Scholar]
20. A. Mehmood, M. Iqbal, Z. Mehmood, A. Irtaza and M. Nawaz, “Prediction of heart disease using deep convolutional neural networks,” Arabian Journal for Science and Engineering, vol. 46, no. 4, pp. 3409–3422, 2021. [Google Scholar]
21. D. Meenakshi-Siddharthan, C. Livia, T. Peterson, P. Stalboerger and Z. Attia, “Artificial intelligence–derived electrocardiogram assessment of cardiac age and molecular markers of senescence in heart failure,” In Mayo Clinic Proceedings, vol. 98, no. 3, pp. 372–385, 2023. [Google Scholar]
22. S. Siddiqui, A. Athar, M. Khan, S. Abbas, Y. Saeed et al., “Modelling, simulation and optimization of diagnosis cardiovascular disease using computational intelligence approaches,” Journal of Medical Imaging and Health Informatics, vol. 10, no. 5, pp. 1005–1022, 2020. [Google Scholar]
23. A. Sau, S. Ibrahim, D. Kramer, J. Waks, N. Qureshi et al., “Artificial intelligence–enabled electrocardiogram to distinguish atrioventricular re-entrant tachycardia from atrioventricular nodal re-entrant tachycardia,” Cardiovascular Digital Health Journal, vol. 4, no. 2, pp. 60–67, 2023. [Google Scholar] [PubMed]
24. P. Farjo, N. Yanamala, N. Kagiyama, H. Patel, G. Casaclang-Verzosa et al., “Prediction of coronary artery calcium scoring from surface electrocardiogram in atherosclerotic cardiovascular disease: A pilot study,” European Heart Journal-Digital Health, vol. 1, no. 1, pp. 51–61, 2020. [Google Scholar] [PubMed]
25. M. Dilli Babu and M. Sambath, “Heart disease prognosis and quick access to medical data record using data lake with deep learning approaches,” International Journal of Intelligent Systems and Applications in Engineering, vol. 11, no. 3s, pp. 292–300, 2023. [Google Scholar]
26. P. Mathur, P. Sharma and K. Veer, “Comparison of soft computing and optimization techniques in classification of ECG signal,” Recent Advances in Computer Science and Communications, vol. 16, no. 2, pp. 37–43, 2023. [Google Scholar]
27. S. Singhal and M. Kumar, “A systematic review on artificial intelligence-based techniques for diagnosis of cardiovascular arrhythmia diseases: Challenges and opportunities,” Archives of Computational Methods in Engineering, vol. 30, no. 2, pp. 865–888, 2023. [Google Scholar]
28. M. Mohan, G. Veena, U. Pavitha and H. Vinod, “Analysis of ECG data to detect sleep apnea using deep learning,” Journal of Survey in Fisheries Sciences, vol. 10, no. 4S, pp. 371–376, 2023. [Google Scholar]
29. S. Briongos-Figuero, A. Estévez-Paniagua, A. Sánchez-Hernández and R. Muñoz-Aguilera, “Combination of current and new electrocardiographic-based criteria: A novel score for the discrimination of left bundle branch capture,” Europace, vol. 25, no. 3, pp. 1051–1059, 2023. [Google Scholar] [PubMed]
30. S. Sarkar, S. Majumder, J. Koehler and S. Landman, “An ensemble of features based deep learning neural network for reduction of inappropriate atrial fibrillation detection in implantable cardiac monitors,” Heart Rhythm O2, vol. 4, no. 1, pp. 51–58, 2023. [Google Scholar]
31. A. Suneetha and T. Mahalngam, “Fine tuning bert based approach for cardiovascular disease diagnosis,” International Journal of Intelligent Systems and Applications in Engineering, vol. 11, no. 6s, pp. 59–66, 2023. [Google Scholar]
32. Y. Chang, M. Dong, B. Wang, M. Ren and L. Fan, “Review of ex vivo cardiac electrical mapping and intelligent labeling of atrial fibrillation substrates,” Chinese Journal of Electrical Engineering, vol. 9, no. 1, pp. 93–103, 2023. [Google Scholar]
33. B. Omarov, A. Tursynova, O. Postolache, K. Gamry, A. Batyrbekov et al., “Modified UNET model for brain stroke lesion segmentation on computed tomography images,” Computers, Materials & Continua, vol. 71, no. 3, pp. 4701–4717, 2022. [Google Scholar]
34. M. Abubaker and B. Babayiğit, “Detection of cardiovascular diseases in ECG images using machine learning and deep learning methods,” IEEE Transactions on Artificial Intelligence, vol. 4, no. 2, pp. 373–382, 2022. [Google Scholar]
35. L. Jing, A. Ulloa Cerna, C. Good, N. Sauers, G. Schneider et al., “A machine learning approach to management of heart failure populations,” Heart Failure, vol. 8, no. 7, pp. 578–587, 2020. [Google Scholar] [PubMed]
36. A. Hannun, P. Rajpurkar, M. Haghpanahi, G. Tison, C. Bourn et al., “Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network,” Nature Medicine, vol. 25, no. 1, pp. 65–69, 2019. [Google Scholar] [PubMed]
37. M. Karri, C. Annavarapu and K. Pedapenki, “A Real-time cardiac arrhythmia classification using hybrid combination of delta modulation 1D-CNN and Blended LSTM,” Neural Processing Letters, vol. 55, no. 2, pp. 1499–1526, 2023. [Google Scholar]
38. E. Prabhakararao and S. Dandapat, “Myocardial infarction severity stages classification from ECG signals using attentional recurrent neural network,” IEEE Sensors Journal, vol. 20, no. 15, pp. 8711–8720, 2020. [Google Scholar]
39. A. Goswami, G. Bhavekar and P. Chafle, “Electrocardiogram signal classification using VGGNet: A neural network based classification model,” International Journal of Information Technology, vol. 15, no. 1, pp. 119–128, 2023. [Google Scholar]
40. M. Wong, H. Hei, S. Lim and E. Ng, “Applied machine learning for blood pressure estimation using a small, real-world electrocardiogram and photoplethysmogram dataset,” Mathematical Biosciences and Engineering, vol. 20, no. 1, pp. 975–997, 2023. [Google Scholar] [PubMed]
41. A. Halim and S. Isa, “Electrocardiogram signal classification for diagnosis sudden cardiac death using 2D CNN and LSTM,” International Journal of Intelligent Systems and Applications in Engineering, vol. 11, no. 4s, pp. 558–564, 2023. [Google Scholar]
42. H. Rai and K. Chatterjee, “Hybrid CNN-LSTM deep learning model and ensemble technique for automatic detection of myocardial infarction using big ECG data,” Applied Intelligence, vol. 52, no. 5, pp. 5366–5384, 2023. [Google Scholar]
43. A. H. Khan and M. Hussain, “ECG images dataset of cardiac patients,” Mendeley Data, Version 2, 2021. [Online]. Available: https://doi.org/10.17632/gwbz3fsgp8.2 [Google Scholar] [CrossRef]
44. G. Moody and R. Mark, “The impact of the MIT-BIH arrhythmia database,” IEEE Engineering in Medicine and Biology Magazine, vol. 20, no. 3, pp. 45–50, 2001. [Google Scholar] [PubMed]
45. A. Goldberger, L. Amaral, L. Glass, J. Hausdorff, P. Ivanov et al., “PhysioBank PhysioToolkit and PhysioNet: Components of a new research resource for complex physiologic signals,” Circulation, vol. 101, no. 23, pp. e215–e220, 2000. [Google Scholar] [PubMed]
46. E. Özbay, A. Çinar and F. Özbay, “3D human activity classification with 3D zernike moment based convolutional, LSTM-deep neural networks,” Traitement du Signal, vol. 38, no. 2, pp. 269–280, 2021. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF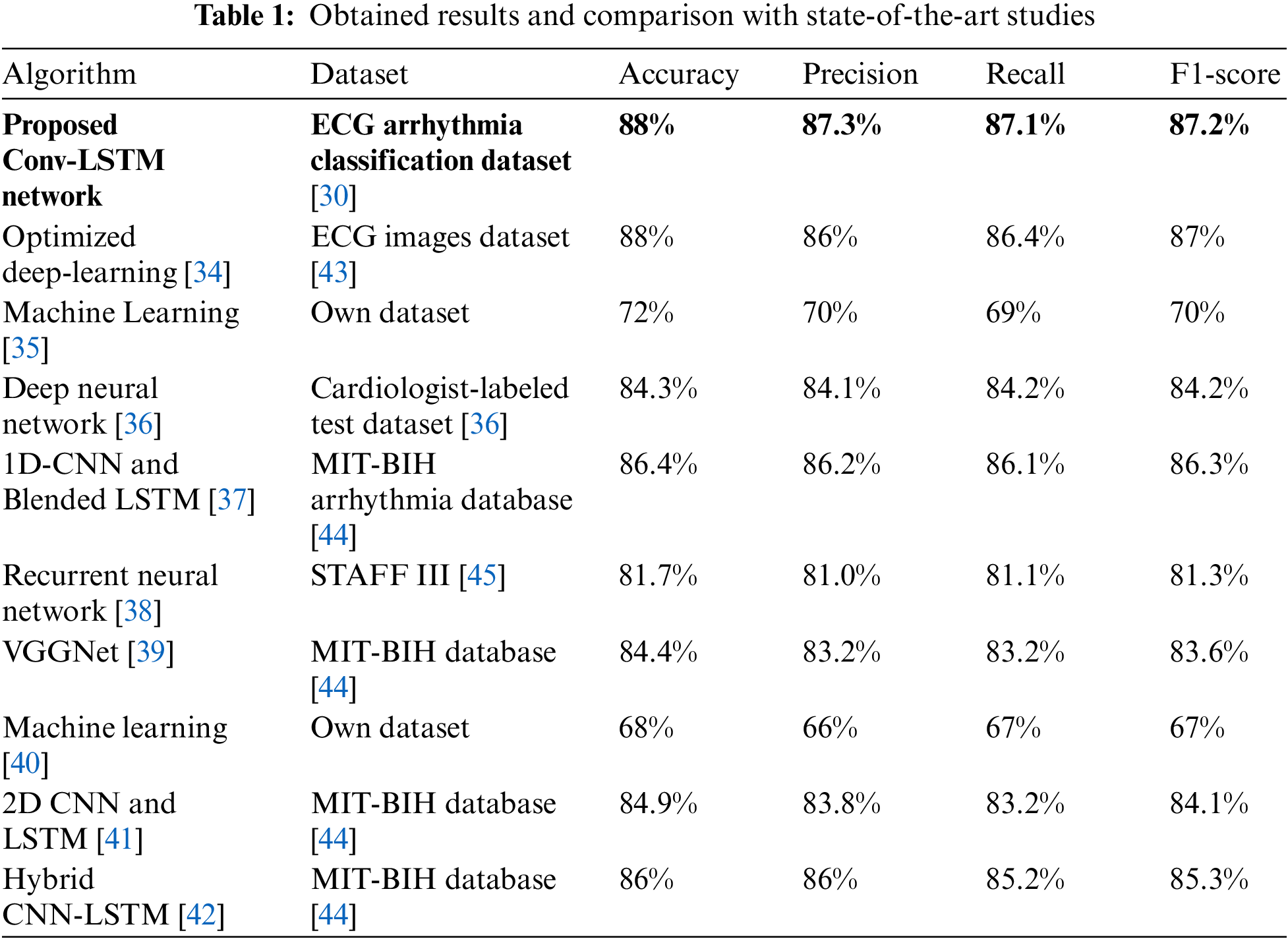
 Downloads
Downloads
 Citation Tools
Citation Tools