 Open Access
Open Access
ARTICLE
Identification and Characterization of ZF-HD Genes in Response to Abscisic Acid and Abiotic Stresses in Maize
1 College of Agronomy and Biotechnology, Southwest University, Chongqing, 400715, China
2 Engineering Research Center of South Upland Agriculture, Ministry of Education, Chongqing, 400715, China
3 Key Laboratory of Application and Safety Control of Genetically Modified Crops, Chongqing, 400715, China
* Corresponding Author: Chaoxian Liu. Email:
# These authors contributed equally to this work
(This article belongs to the Special Issue: Identification of Genetic/Epigenetic Components Responding to Biotic and Abiotic Stresses in Crops)
Phyton-International Journal of Experimental Botany 2023, 92(3), 707-723. https://doi.org/10.32604/phyton.2023.024338
Received 29 May 2022; Accepted 19 August 2022; Issue published 29 November 2022
Abstract
The zinc finger homeodomain (ZF-HD) genes belong to the homeobox gene family, playing critical roles in flower development and stress response. Despite their importance, however, to date there has been no genome-wide identification and characterization of the ZF-HD genes that are probably involved in stress responses in maize. In this study, 24 ZF-HD genes were identified, and their chromosomal locations, protein properties, duplication patterns, structures, conserved motifs and expression patterns were investigated. The results revealed that the ZF-HD genes are unevenly distributed on nine chromosomes and that most of these genes lack introns. Six and two ZF-HD genes have undergone segmental and tandem duplication, respectively, during genome expansion. These 24 ZF-HD transcription factors were classified into six major groups on the basis of protein molecular evolutionary relationship. The expression profiles of these genes in different tissues were evaluated, resulting in producing two distinct clusters. ZF-HD genes are preferentially expressed in reproductive tissues. Furthermore, expression profiles of the 24 ZF-HD genes in response to different kinds of stresses revealed that ten genes were simultaneously up-regulated under ABA, salt and PEG treatments; meanwhile four genes were simultaneously down-regulated. These findings will pave the way for deciphering the function and mechanism of ZF-HD genes on how to implicate in abiotic stress.Keywords
ZF-HD transcription factors (TFs) play critical roles in regulating flower development and stress response [1–3]. These proteins contain two highly conserved zinc finger motifs, the upstream region of which is termed the zinc finger-homeodomain (ZF-HD). The ZF-HD proteins have a C-terminal domain in which resides a DNA binding homeodomain and an N-terminal domain that contains five conserved cysteine residues and at least three conserved histidine residues for potential zinc binding [4]. The ZF domain not only participates in DNA binding but also can enhance protein-DNA interactions, which are mediated by the HD domain [5,6].
ZF-HD genes were first identified in the C4 plant Flaveria trinervia [7]. Subsequently, a growing number of ZF-HD family members have been identified in many plants such as Arabidopsis thaliana [8], Oryza sativa [9], cucumber [10], Vitis vinifera [11] and wheat [12]. Previous studies have confirmed that ZF-HD genes play critical roles in plant growth and development. In Arabidopsis, the identified 14 ZF-HD genes were all florally expressed, and the loss-of-function mutations for six genes individually showed no obvious phenotypes, indicating those genes play overlapping regulatory roles in floral development [8]. SlZHD17, functioning in chlorophyll and carotenoid metabolism, have a pleiotropic effect on tomato development; the knockout plants showed dwarfism, accelerated flowering, earlier fruit harvest, as well as larger chloroplasts and higher chlorophyll content [13]. Many studies have also demonstrated that ZF-HD genes play essential roles in the responses to abiotic stress. For example, the expressions of ZF-HD TFs are highly induced by abiotic stress in Chinese cabbage and wheat [14,15]. In rice, four ZF-HD TFs that bind to the OsDREB1B promoter are induced by low temperatures, drought, and mechanical stresses [16]. In addition, ZF-HD proteins have been demonstrated to function in the establishment of expression patterns of the C4 phosphoenolpyruvate carboxylase gene in F. trinervia [7], and play critical roles in regulating rice leaf curling by influencing the formation and distribution of bulliform cells [17].
Although WOX, HD-ZIP and TALE subfamilies of HD genes have been identified in maize [18–20], to date there have been no comprehensive analyses of ZF-HD genes in this important crop plant. The completion of maize genome sequencing and recent developments in bioinformatics methods now make it possible to characterize the maize ZF-HD family on a genome-wide scale. In this study, we identified ZF-HD transcription factors in the maize genome, and performed phylogenetic analysis and classification to explore the evolutionary relationship among members of the ZF-HD gene family. We also describe features of ZF-HD gene structure and conserved motifs, along with expression profiling of these in different maize organs and in response to different abiotic stresses. This study will lay a solid foundation for elucidating the function the ZF-HD TFs in response to abiotic stresses in maize.
2.1 Identification of ZF-HD Genes in Maize and Phylogenetic Tree Construction of ZF-HD Proteins from Maize, Rice and Arabidopsis
To identify ZF-HD genes in maize, the maize genome sequence was downloaded from MaizeGDB (http://www.maizegdb.org), and a local database based on the nucleotide and protein sequences was constructed. The Hidden Markov Model (HMM) profile of ZF-HD dimer domains was obtained using HMMER (an online web-based software for biological sequence analysis), and used as a search query to identify possible maize ZF-HD TFs using DNA tools [21,22]. All candidate sequences were confirmed using Pfam (http://pfam.xfam.org/family/PF04770) and InterPro (http://www.ebi.ac.uk/interpro/entry/IPR006456). Sequences of the ZF-HD genes of Arabidopsis and rice were downloaded from the TFDB database (http://planttfdb.cbi.pku.edu.cn/). Finally, all ZF-HD TFs were named in accordance with the order of genes on maize chromosomes. In order to determine the phylogenetic relationships among ZF-HD proteins, all protein sequences of ZF-HD in maize, rice, and Arabidopsis were aligned using MAFFTV_7 (http://mafft.cbrc.jp/alignment/server/), and then a phylogenetic tree was constructed using the neighbor-joining (N-J) method in MEGA v5.0 [23]. For statistical reliability, bootstrap test was set as 1,000 replicates to evaluate the significance of each node.
2.2 Chromosomal Localization, Protein Properties and Duplications
The details of ZF-HD gene family members in maize, rice, and Arabidopsis, including chromosomal location, amino acid sequence, and coding sequence (CDS), were obtained from MaizeGDB (https://www.maizegdb.org/) and Gramene (http://ensembl.gramene.org/). The molecular weight (Mw), theoretical isoelectric point (pI), and protein length were calculated using ExPASy (http://web.expasy.org/protparam/). The subcellular localization of ZF-HD proteins was predicted using CELLO v.2.5 (https://doi.org/10.1371/journal.pone.0099368), and a chromosome location image of maize ZF-HD genes was generated using MapInspect (http://mapinspect.software.informer.com/). Maize ZF-HD gene duplication events were investigated according to the previous report by Wei et al. [24]. Paralogs that mapped within the same chromosomal block were considered to be segmental duplications, whereas paralogs separated by a maximum of five genes were considered to be tandem duplications.
2.3 Structural Analysis of Maize ZF-HD Genes and Domain Prediction
The exon-intron structures of ZF-HD genes were analyzed using the Gene Structure Display Server using both genomic DNA and the corresponding CDS sequences (http://gsds.gao-lab.org/). The conserved motifs of maize ZF-HD genes were analyzed using MEME (http://meme-suite.org/tools/meme) with the following parameters: 10–1,000 amino acids were adopted as the optimum width, and any number of repetitions of a motif and maximum number of motifs were set as 15, respectively. MEME motifs were annotated using the Interpro program.
2.4 The Expression Pattern of Maize ZF-HD Genes
To examine the expression patterns of the 24 identified maize ZF-HD genes in different tissues, we collected expression profile data from qTeller (https://qteller.maizegdb.org/), including that for whole anthers, pollen, mature silk, mature leaf, seedling roots, embryo at 14 days after pollination (DAP), endosperm at 14 DAP, ovaries 1 DAP, ear primordia, tassel primordia, P7 ligule and P7 sheath (P indicates plastochron number). We then employed MEV (https://sourceforge.net/projects/mev-tm4/) to perform cluster analysis of maize ZF-HD gene expression. PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was used to predict cis-elements in the 2,000-bp promoter region of the 24 maize ZF-HD genes [25].
2.5 Plant Materials and Stress Treatments
The B73 inbred line was grown in a greenhouse with a 14-h light and 10-h dark cycle at 28°C–30°C. For stress and hormone treatments, 10-day-old maize seedlings were firstly cultured in modified Hoagland solution for two days, and then treated in the solution plus 200 mM NaCl, 15% (w/v) PEG and 100 µM ABA, respectively. The seedling roots were sampled at 0, 1, 2, 5, and 10 h after treatment. Each treatment had three biological replicates.
2.6 RNA Isolation and Quantitative Real-Time PCR (qRT-PCR) Analyses
Total RNA was extracted from all collected samples using RNAprep pure Plant Kisst (Tiangen, China), and then the first strand of cDNA was synthesized using RevertAid First Strand cDNA Synthesis Kit (Thermo, USA). Gene specific primers were designed using Primer Express 3.0 (Applied Biosystems, USA). Each reaction was performed in 12 μL reaction mixture containing 2 μL of diluted cDNA sample, 9.2 μL of 2 × Power SYBR Green PCR Master Mix (Applied Biosystems, USA), and 0.8 μL each of forward and reverse gene-specific primers. The thermal cycle used was as follows: 95°C for 3 min, followed by 40 cycles at 95°C for 10 s, and 64°C for 30 s. In this study, we performed three technical replicates for each gene. The relative expression levels of each candidate gene were calculated using the 2–ΔΔCt method [26], and the no stress treatment (0 h) was normalized to 1.
3.1 Characterization and Chromosomal Localization of ZF-HD Genes in Maize
The amino acid sequence of the ZF-HD dimer domain was used as a query to identify maize ZF-HD proteins from a local database, the candidate proteins lacking a ZF-HD domain were removed using the Pfam and InterPor databases. A total of 24 ZF-HD genes were accordingly detected in the maize genome. The predicted ZF-HD genes were named based on gene order on the chromosomes (Table 1).
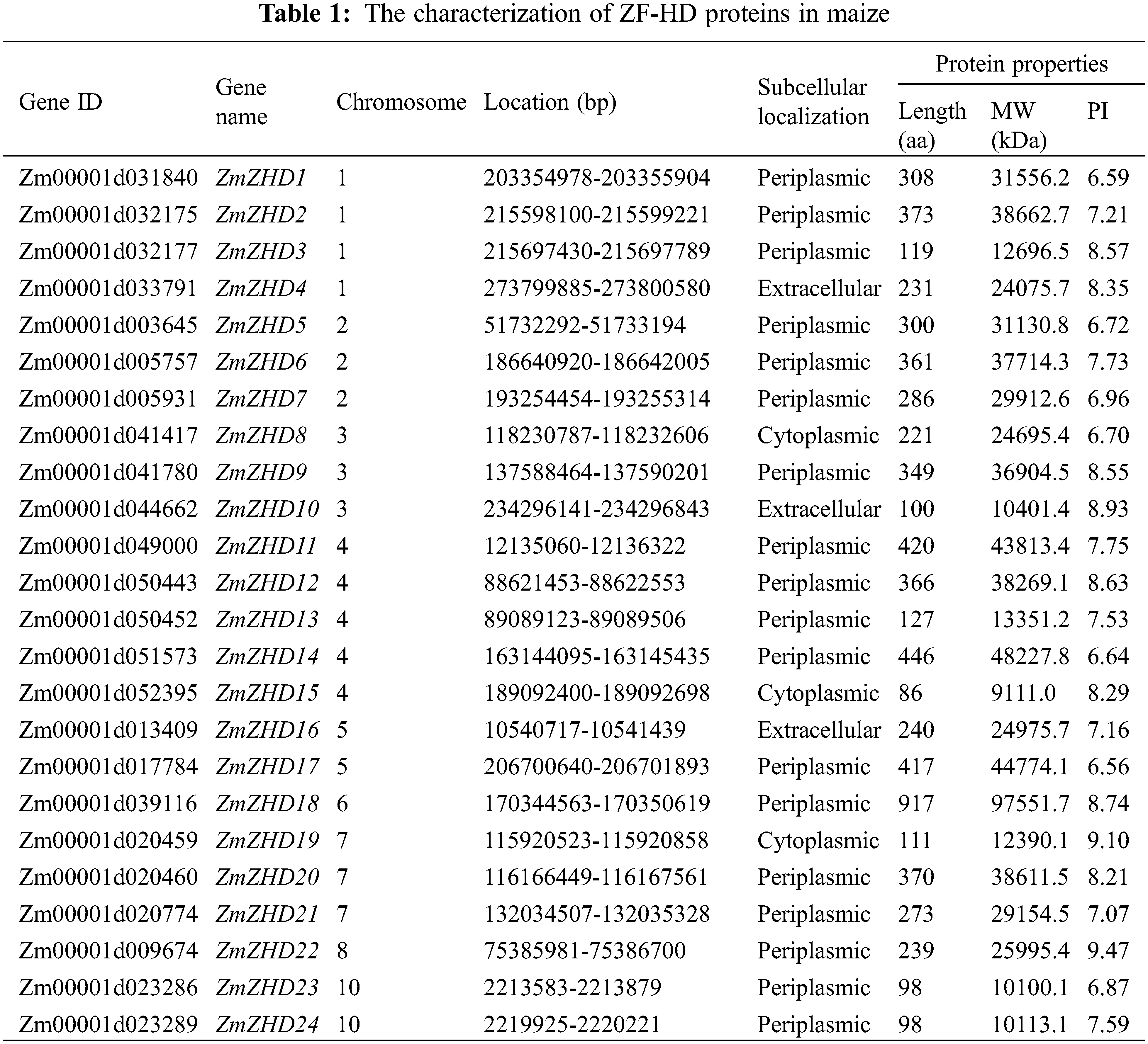
The identified maize ZF-HD genes encoded proteins with lengths ranging from 86 to 917 amino acids, Mw from 9,111 to 97,551.7 kDa, and pI from 6.56 to 9.47. The distribution of the 24 maize ZF-HD genes on nine of the ten maize chromosomes was uneven (Fig. 1). Chromosome 4 harbored five ZF-HD genes, whereas only one gene was detected on each of chromosomes 6 and 8. Among the other genes, four were located on chromosome 1, three each on chromosomes 2, 3, and 7, and two each on chromosomes 5 and 10. Phylogenetic analysis of the maize ZF-HD genes revealed eight sister pairs showing a close relationship, namely, ZmZHD7/21, ZmZHD14/17, ZmZHD22/18, ZmZHD4/16, ZmZHD13/3, ZmZHD12/2, ZmZHD6/20, and ZmZHD23/24 (Fig. 2). Three pairs, ZmZHD14/17, ZmZHD22/18 and ZmZHD4/16, detected in the same duplicated chromosomal blocks, have undergone segmental duplications. Intriguingly, gene pair ZmZHD23/ZmZHD24, was located in the same duplicated chromosomal blocks, and the physical distance between these two genes was only 6.3 kb, which indicated that this gene cluster has undergone tandem duplication. It is noteworthy that three of the sister pairs (ZmZHD13/3, ZmZHD12/2, and ZmZHD6/20) were found to be located in very close proximity to the duplicated chromosomal blocks. Moreover, they had similar gene structures and a close evolutionary relationship, indicating that they are putative segmentally duplicated genes. These findings tended to indicate that segmental duplication is an important mechanism in maize ZF-HD gene family expansion.
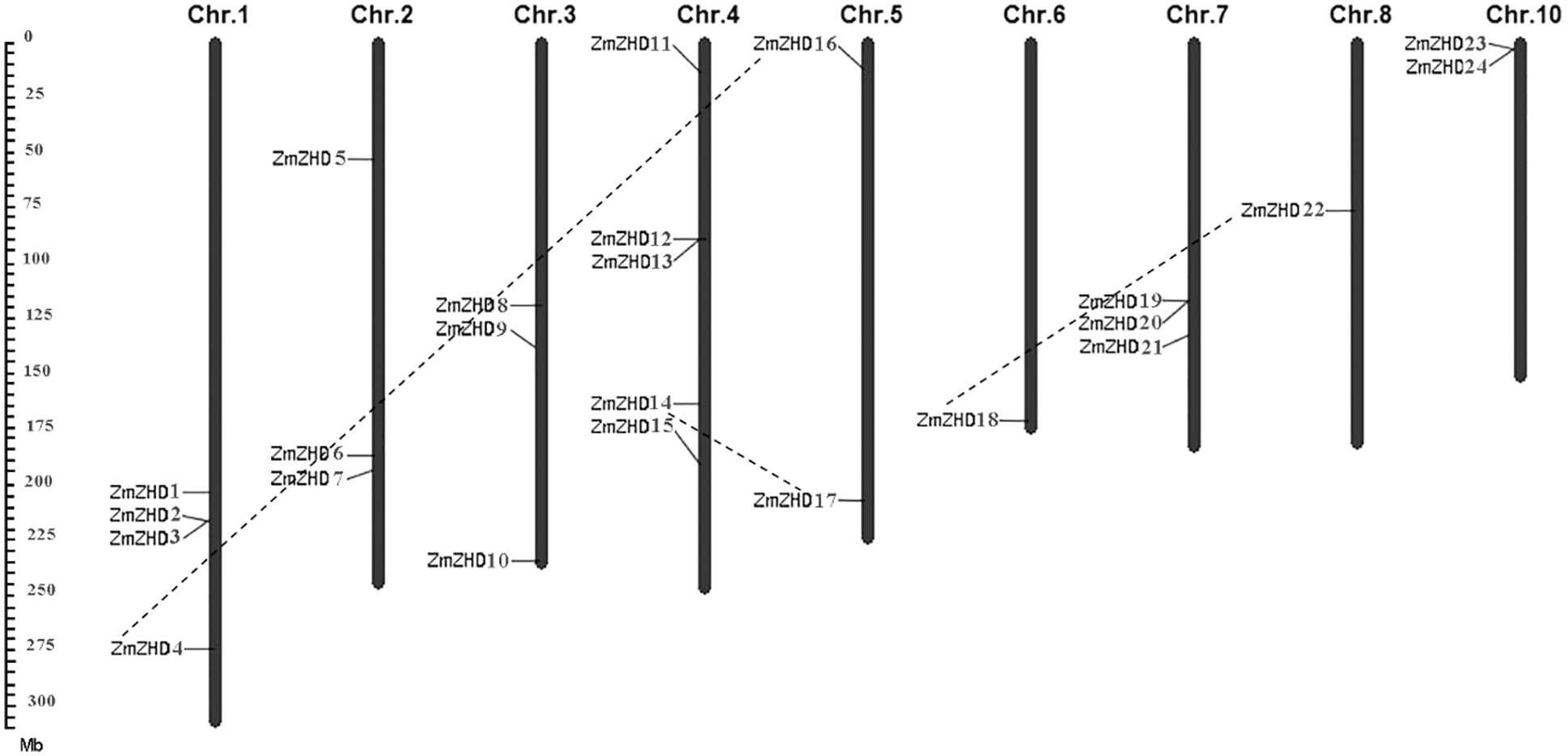
Figure 1: Distribution of maize ZF-HD family members on maize chromosomes. The chromosome numbers are indicated at the top of each bar. The segmentally duplicated genes are connected by dash lines
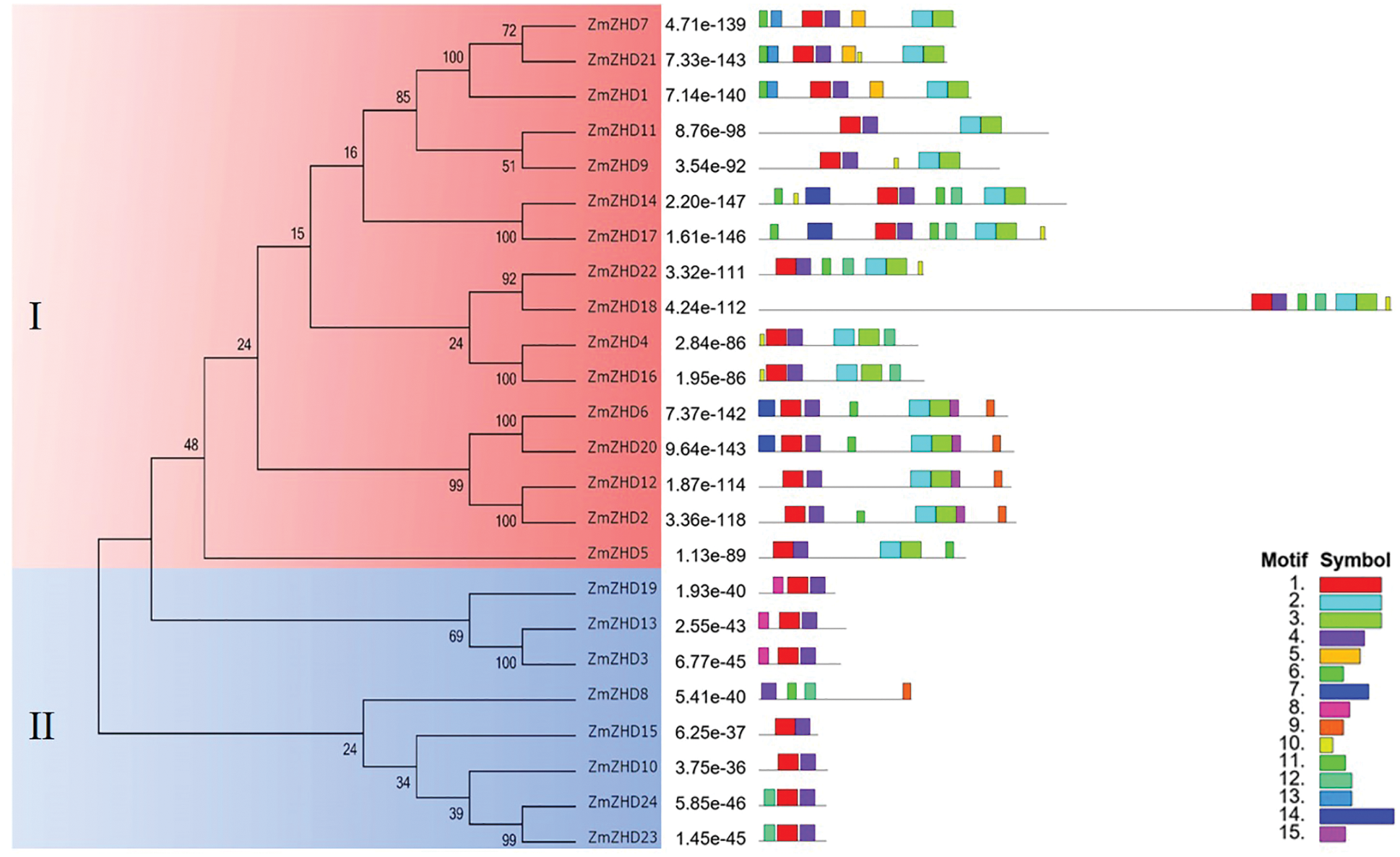
Figure 2: Phylogenetic analysis and conserved motif distribution of maize ZF-HD proteins. The constructed phylogenetic tree is shown on the left side of the figure. The legends on the bottom right corner of the figure represent different motifs
3.2 Identification of Conserved Motifs in Maize ZF-HD Proteins
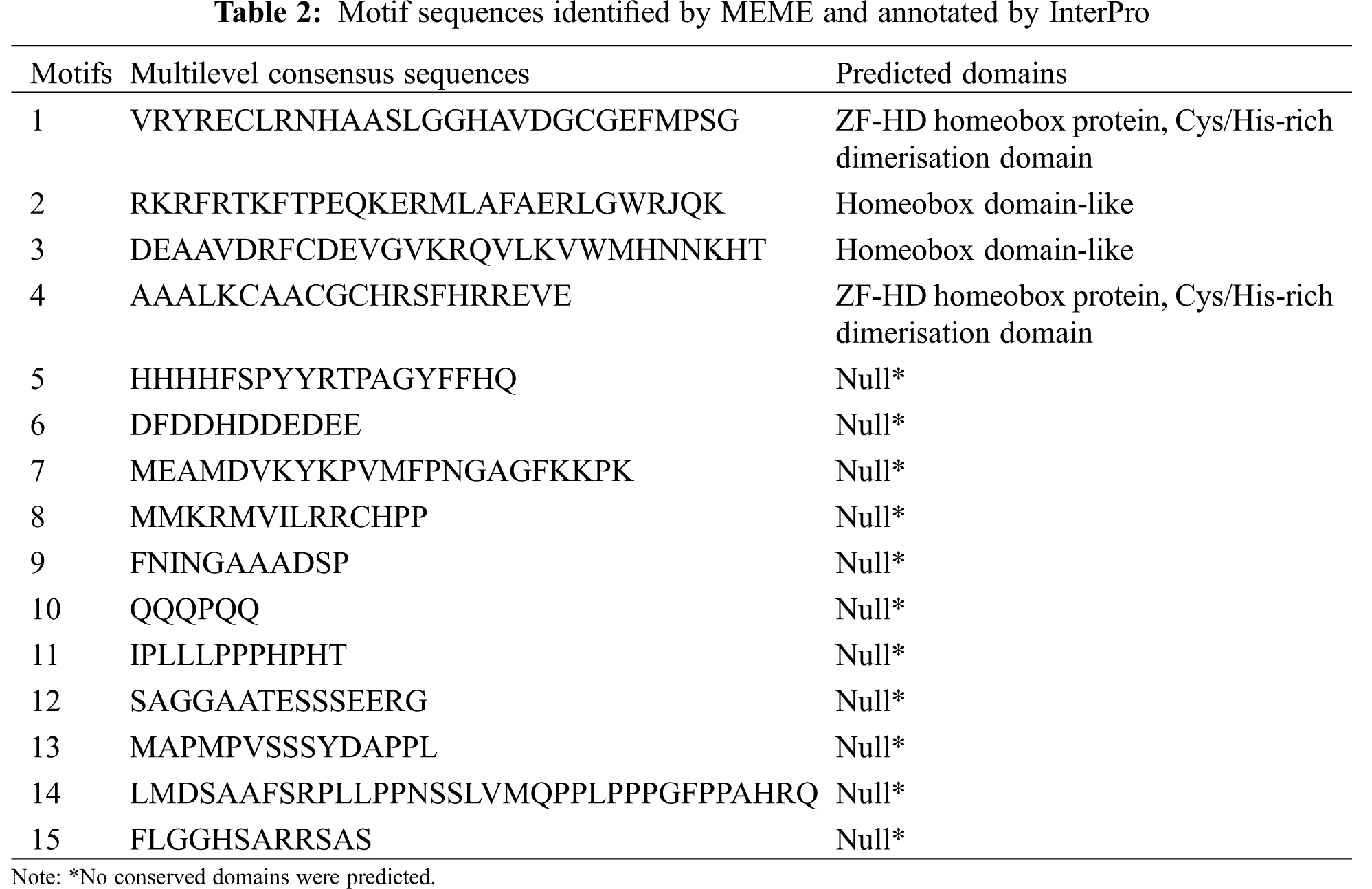
In order to investigate the sequence feature of maize ZF-HD proteins, the conserved motifs of these proteins were obtained and annotated using MEME and Interpro, respectively. As a result, 15 motifs were identified in the maize ZF-HD proteins (Fig. 2, Table 2). The motif distribution was consistent with protein phylogenetic evolution, with members in the same clade generally sharing similar motifs. On the basis of motif distribution, the maize ZF-HD proteins were divided into two clades (Fig. 2). Almost all of the maize ZF-HD proteins harbored motifs 1 and 4 (the exception being ZmZHD8, which had only motif 4). Sixteen (Clade I) out of the 24 ZF-HD proteins shared four common motifs (motifs 1, 2, 3, and 4); seven proteins (Clade II), lacking motifs 2 and 3, just contained motifs 1 and 4. Motifs 2 and 3, which were located close to the C terminus, represented homeobox domain-like structures that are involved in DNA binding in the transcriptional regulation of target genes. Motifs 1 and 4 represented an indispensable basic conserved ZF-HD domain (Cys/His-rich dimerization domain), which is sufficient to confer homo- or heterodimer formation between proteins and plays a critical role in protein functions [7]. The motifs 5–15 were mainly distributed in Clade I apart from motifs 1–4, whereas only motif 8, motif 9, motif 11 and motif 12 resided in Clade II proteins. In brief, the distribution of motifs in Clades I and II indicated the functional divergence of ZF-HD gene in maize.
3.3 ZF-HD Phylogenetic and Gene Structure Analysis
To analyze the evolutionary history of the ZF-HD TF family, we performed a molecular phylogeny analysis on the 24 maize ZF-HD protein sequences, in conjunction with 15 from rice and 17 from Arabidopsis (Tables S1 and S2). We accordingly found that these 56 ZF-HD genes could be clustered into six major groups: groups A, B, C, D, E and F, containing 15, 3, 8, 13, 5 and 12 genes, respectively (Fig. 3). A notable feature of the constructed phylogenetic tree was that almost all the Arabidopsis ZF-HD genes were clustered together, whereas the ZF-HD genes from rice and maize were clustered together. In addition, many of the maize and rice ZF-HD genes fell into orthologous pairs, including ZmZHD1/OsZHD8, ZmZHD11/OsZHD13, ZmZHD9/OsZHD15, ZmZHD5/OsZHD4, and ZmZHD19/OsZHD9, which indicated that these ZF-HD orthologous genes had conserved functions, even though the maize and rice genomes undergone markedly different recombination and replication events subsequent to their divergence from a common ancestor. We also performed structural analyses of the ZF-HD genes and found that 46 out of 56 ZF-HD genes had no introns, which contrasts with the structure of other homeobox genes. Six of the genes contained a single intron (two genes each in maize, rice, and Arabidopsis). The general absence of introns in these genes tended to indicate that the structure and functions of these genes were highly conserved, as there was no alternative splicing in these genes.
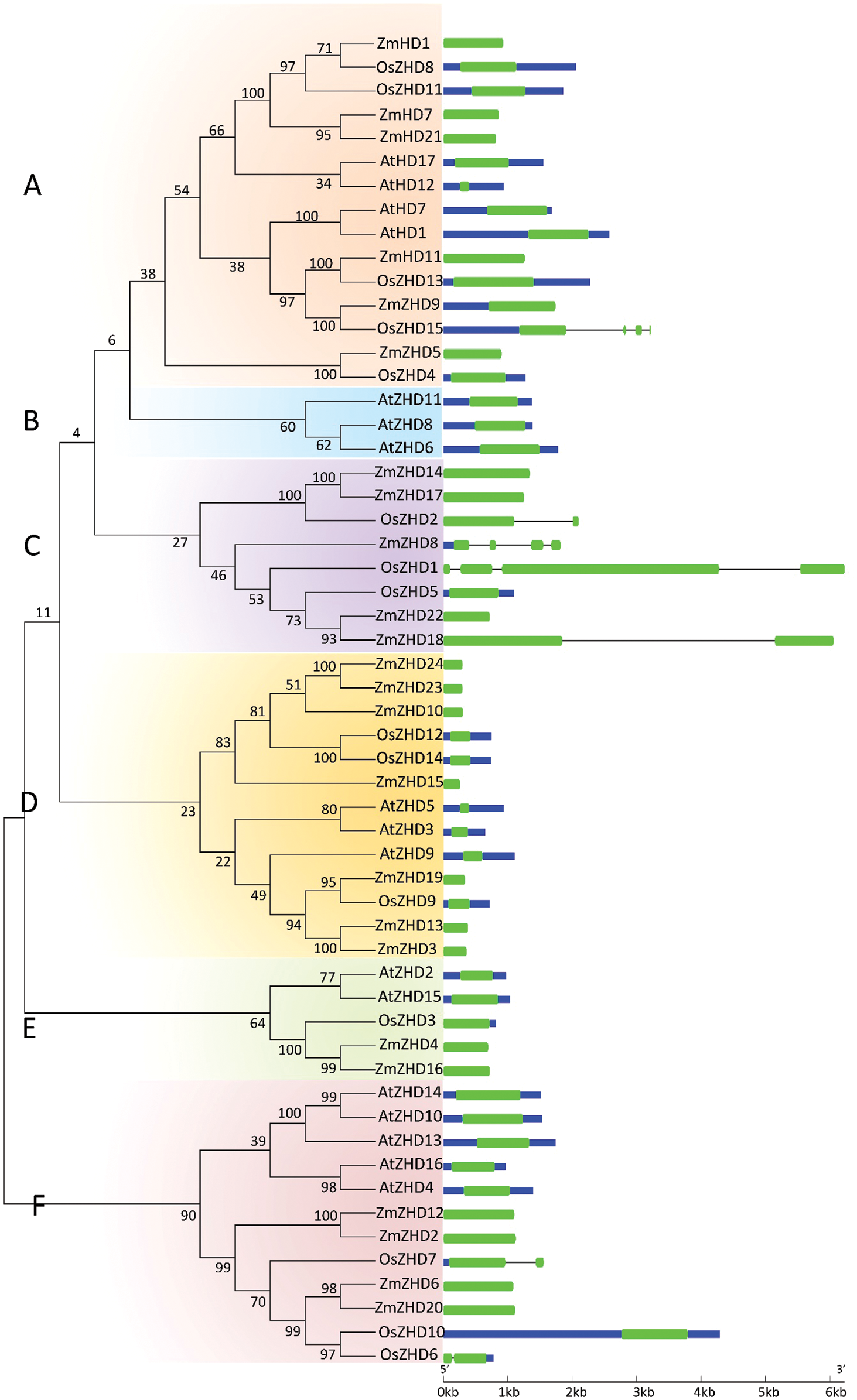
Figure 3: Phylogenetic relationship and gene structure analyses of the ZF-HD protein family in maize, rice, and Arabidopsis. Numbers above or below branches of the tree represent bootstrap values. A, B, C, D, E and F represent the different groups. Exons, introns, and upstream/downstream sequences are represented by green boxes, black lines, and blue boxes, respectively. The colored boxes and black lines are scaled based on the length of genes (the short introns in ZmZHD9 and AtZHD1/11 are not shown in the figure)
3.4 Expression Pattern of Paize ZF-HD Genes in Different Tissues
To explore the expression patterns of maize ZF-HD genes and obtain information for functional analyses, we investigated the expression of the 24 identified maize ZF-HD genes. The relative expression level of these ZF-HD genes was detected in 12 different tissues (Fig. 4). On the basis of cluster analysis, the maize ZF-HD genes were distinctly classified into two major clusters. Cluster A contained 16 genes (ZmZHD4, 1, 12, 2, 5, 9, 17, 6, 11, 20, 18, 16, 14, 7, 21 and 22), and cluster B contained eight genes (ZmZHD8, 10, 19, 3, 13, 15, 23 and 24). The ZF-HD genes in different clusters exhibited distinct expression patterns, but were all preferentially expressed in reproductive tissues. Most genes in cluster A were dominantly expressed in embryo, ear and tassel primordia, half of which were highly expressed in ovary, indicating that these genes probably play regulatory roles in floral and kernel development. Cluster B genes were highly expressed in anther, pollen, mature silk, mature leaf and seedling roots, moderately expressed in embryo, endosperm, ear and tassel primordia. Interestingly, most of ZF-HD genes showed a moderate expression in ligule and sheath. Additionally, three gene pairs, ZmZHD7/21, ZmZHD13/3 and ZmZHD23/24, had a high co-expression relationship consistent with the phylogenetic analysis, indicating that these genes might have conserved roles during growth and development in maize.
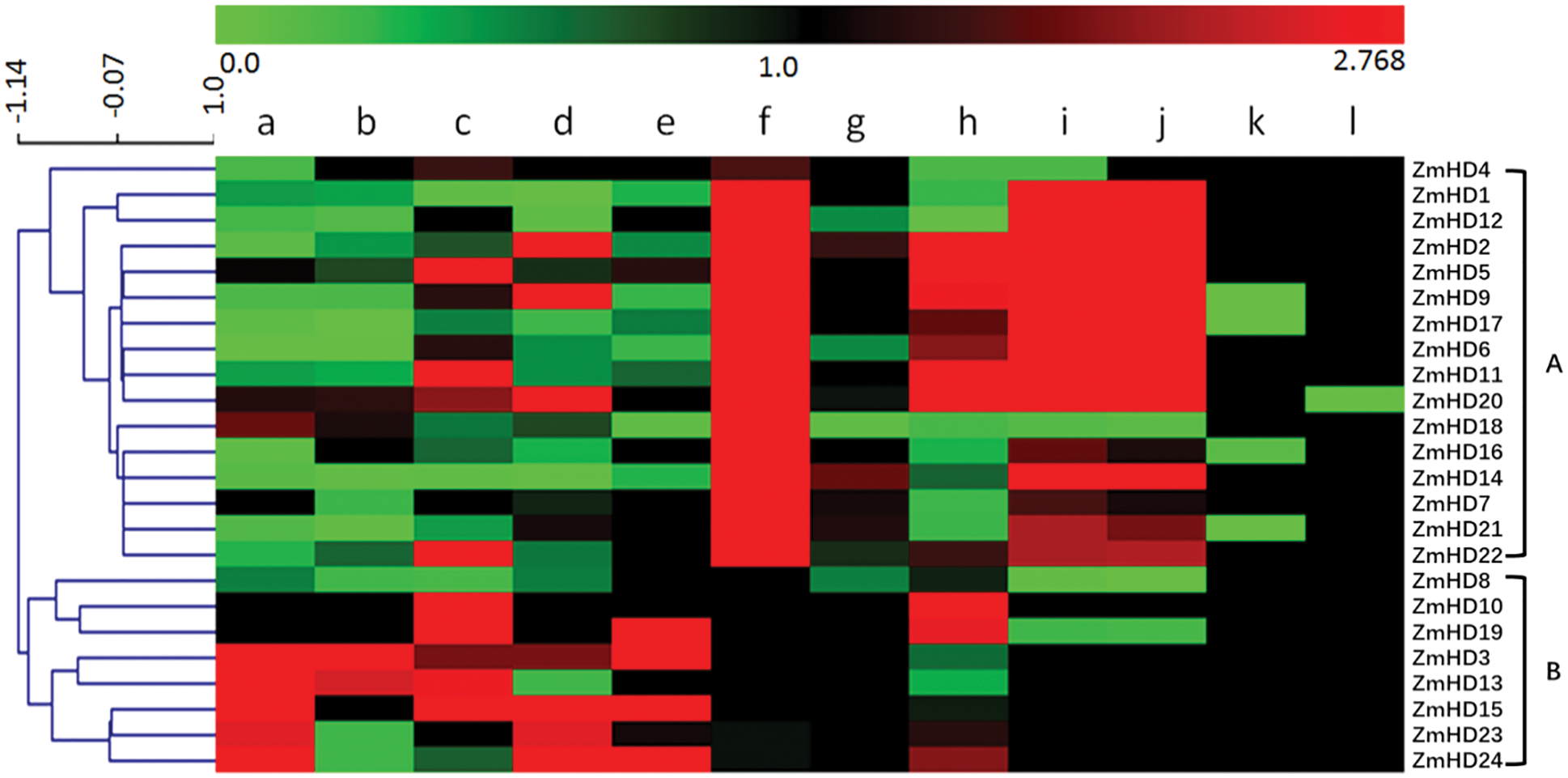
Figure 4: The expression profiles of ZF-HD genes in maize. Red and green color indicates genes with high and low expression levels, respectively. The gradual change in color from green to red indicates the change of gene expression level from low to high. a: whole anthers; b: pollen; c: mature silk; d: mature leaf; e: seedling roots; f: embryo 14 DAP; g: endosperm 14 DAP; h: ovaries 1 DAP; i: ear primordia; j: tassel primordia; k: P7 ligule and l: P7 sheath
3.5 Expression Patterns of ZF-HD Genes in Response to Abiotic Stresses and ABA
By investigating cis-elements in the promoter regions of the 24 maize ZF-HD genes, we found that 18 of these genes contained an ABA-responsive element (ABRE) (Table S3), thereby indicating that they might be involved in ABA signal transduction. This observation prompted us to investigate the expression pattern of the 24 identified maize ZF-HD genes in response to ABA, high salt, and drought treatments. Gene specific primers were designed (Table S4) and gene expression patterns under different conditions were detected. Data analysis revealed that the expression of 16 genes was obviously up-regulated under ABA treatment, 15 genes under salt treatment and 14 genes under PEG treatment (Fig. 5, Table S5), among which ten genes (ZHD1, ZHD5, ZHD6, ZHD7, ZHD8, ZHD11, ZHD12, ZHD18, ZHD19 and ZHD20) were all up-regulated in the three treatments. In addition, we also found that six genes were down-regulated under the ABA and PEG treatments, seven genes under the salt treatment; four genes (ZHD4, ZHD14, ZHD15 and ZHD17) were down-regulated in all the treatments. The results suggested that those genes up- and down-regulated probably were implicated in the abiotic stress.
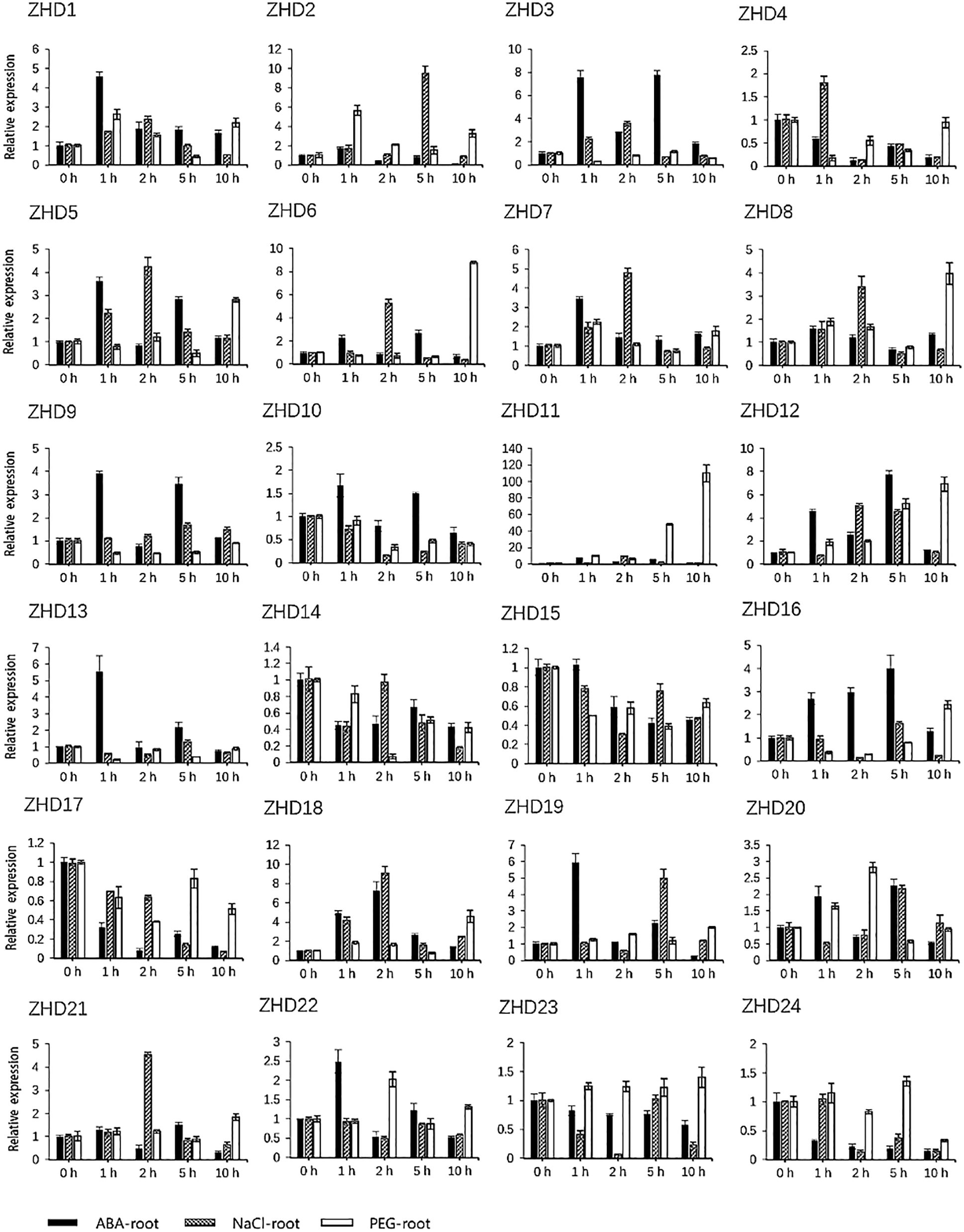
Figure 5: qRT-PCR analysis of the relative expression of ZF-HD genes in response to abiotic stress and ABA treatments. Histograms are the mean ± S.D of n = 3
ZF-HD genes play critical roles in plant development. In the present study, we detected 24 ZF-HD genes in the maize genome, based on a comprehensive method of ZF-HD gene mining. The maize genome is nearly six times larger than the rice genome and 20 times larger than that of Arabidopsis [27], although the number of maize genes is similar to that of rice [28] and only 1.6-fold larger than that of Arabidopsis [29]. However, we found that the number of ZF-HD genes in maize (24) is considerably higher than that in both rice (15) and Arabidopsis (17). Our comparative analysis of the phylogenetic relationships among the ZF-HD genes of maize, rice and Arabidopsis indicated the frequent occurrence of orthologous pairs among rice and maize, notably in a subfamily-specific group (group C) comprising only maize and rice genes, indicating that these orthologous pairs diverged from a common ancestor before the separate of maize and rice and evolved independently in monocot plants. The data thus indicate that maize ZF-HD genes have been more closely related with rice ZF-HD genes than with those of Arabidopsis during the course of plant evolution.
Gene duplication events are known to make a substantial contribution to genome expansion [30], and maize underwent genome duplication events during its early evolution [31,32]. It has been reported that such genome duplication might be a major force in the expansion of ZF-HD genes in angiosperm species [33]. The most compelling evidence for this possibility comes from the identification of four sister pairs of close paralogs that probably underwent tandem or segmental duplications, so genome duplication might have played a prominent role in the expansion of ZF-HD gene number in maize. The expression patterns of ZF-HD genes in maize revealed that four paralogous gene pairs showed similar expression patterns, indicating that they have been highly conserved and showed functional redundancy during long-term evolution. Gene redundancy is prevailing in plant [34,35]. Duplicated paralogous genes have three main evolutionary fates over long periods of evolution, namely, nonfunctionalization, subfunctionalization, or neofunctionalization [36,37]. By comparing and analyzing the expression patterns of the four paralogous gene pairs identified in the present study, we found that some showed different expression patterns under a specific treatment. For example, the gene pair ZmZHD4 and ZmZHD16 showed different expression patterns in the drought treatment (Fig. 5), indicating that duplicated genes might have undergone neofunctionalization during the course of evolution.
ZF-HD genes in plant could be classified into eight clades based on gene structural features and motif distribution [38]. In the study, the phylogenetic tree of 56 ZF-HD proteins in maize, rice and Arabidopsis consisted of six major groups (Fig. 3), and genes within the same group showed similar structures and motifs. Moreover, we also found that most of the ZF-HD genes in maize lacked introns, which is ubiquitous in plant [8,12,14,39]. It is well-known that intronless genes cannot undergo alternative splicing, and consequently they have maintained a highly conserved gene structure and relatively fixed function during evolutionary history [40,41]. Our investigation of the expression patterns of ZF-HD genes in 12 different maize tissues discovered that about half of the identified maize ZF-HD genes were preferentially expressed in the tassel and ear, suggesting that these ZF-HD genes play overlapping regulatory roles in maize floral development as well as Arabidopsis floral development [8].
Maize is a staple crop for food, animal feed and industrial raw materials. The production of maize is, however, threatened to varying degrees by different abiotic stresses, such as drought and salinity [42]. As terrestrial plant-specific TFs, members of the ZF-HD gene family play a regulatory role under abiotic stress [43–46]. Our results revealed that most of maize ZF-HD genes under drought, salt and ABA environments responded to stresses. We observed that numerous genes’ expression was greatly induced, as many as ten genes including ZmZHD1, ZmZHD5 and ZmZHD11 (Table S5) were all up-regulated in the three treatments. OsZHD4 (Os04g35500), OsZHD8 (Os08g37400) and OsZHD13 (Os11g13930), the close homologs of ZmZHD5, ZmZHD1 and ZmZHD11, respectively, were confirmed to be differentially regulated by different abiotic stress conditions [16]. In addition, we also found some genes were all down-regulated in treatments, which may act as a negative regulator. Although the function of most of ZF-HD genes in maize has not yet been elucidated, it is not difficult to reach a conclusion that these ZF-HD genes probably are key players under abiotic stress.
In this study, 24 ZF-HD genes were identified in maize, and their chromosome location, protein characteristic, replication mode, gene structure, conserved motif, expression mode and protein molecular evolution were analyzed. In addition, the expression profiles of these genes in different tissues and different abiotic stresses were evaluated. The results revealed that most of ZF-HD genes in maize showed a significant response to abiotic stress. Our results provide valuable clues for deciphering the roles of ZF-HD genes under abiotic stress.
Compliance with Ethical Standards: The authors declare that the review is in compliance with ethical standards of the journal.
Research Involving Human Participants and/or Animals: The authors declare that the manuscript does not contain research involving Human Participants and/or Animals.
Authorship: The authors confirm their contribution to the paper as follows: Chaoxian Liu conceived and designed the experiments, Xiaojie Jing and Chunyan Li performed all of the experiments, analyzed the results and wrote the paper; Chengjuan Luo, Chaonan Yao, Jiahao Zhang, Tingting Zhu and Jiuguang Wang participated in partial work in this study.
Funding Statement: This work was supported by Importing, Cultivation and Production for Special Maize (2020LYXZ032).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Abu-Romman, S., Al-Hadid, K. (2017). Novel zinc finger-homeodomain gene from barley (HvZFHD1) is differentially regulated during spike development and under hormonal treatments and abiotic stresses. South African Journal of Botany, 91, 32–36. DOI 10.1016/j.sajb.2013.11.014. [Google Scholar] [CrossRef]
2. Sun, J., Xie, M., Li, X., Li, Z., Wang, Q. et al. (2021). Systematic investigations of the ZF-HD gene family in tobacco reveal their multiple roles in abiotic stresses. Agronomy, 11(3), 406. DOI 10.3390/agronomy11030406. [Google Scholar] [CrossRef]
3. He, K., Li, C., Zhang, Z., Zhan, L., Cong, C. et al. (2022). Genome-wide investigation of the ZF-HD gene family in two varieties of alfalfa (Medicago sativa L.) and its expression pattern under alkaline stress. BMC Genomics, 23(1), 1–14. DOI 10.1186/s12864-022-08309-x. [Google Scholar] [CrossRef]
4. Tran, L. S., Nakashima, K., Sakuma, Y., Osakabe, Y., Qin, F. et al. (2007). Co-expression of the stress-inducible zinc finger homeodomain ZFHD1 and NAC transcription factors enhances expression of the ERD1 gene in Arabidopsis. The Plant Journal, 49(1), 46–63. DOI 10.1111/j.1365-313X.2006.02932.x. [Google Scholar] [CrossRef]
5. Mackay, J. P., Crossley, M. (1998). Zinc fingers are sticking together. Trends in Biochemical Sciences, 23(1), 1–4. DOI 10.1016/S0968-0004(97)01168-7. [Google Scholar] [CrossRef]
6. Hu, W., dePamphilis, C. W., Ma, H. (2008). Phylogenetic analysis of the plant-specific zinc finger-homeobox and mini zinc finger gene families. Journal of Integrative Plant Biology, 50(8), 1031–1045. DOI 10.1111/j.1744-7909.2008.00681.x. [Google Scholar] [CrossRef]
7. Windhövel, A., Hein, I., Dabrowa, R., Stockhaus, J. (2001). Characterization of a novel class of plant homeodomain proteins that bind to the C4 phosphoenolpyruvate carboxylase gene of Flaveria trinervia. Plant Molecular Biology, 45(2), 201–214. DOI 10.1023/A:1006450005648. [Google Scholar] [CrossRef]
8. Tan, Q. K. G., Irish, V. F. (2006). The Arabidopsis zinc finger-homeodomain genes encode proteins with unique biochemical properties that are coordinately expressed during floral development. Plant Physiology, 140(3), 1095–1108. DOI 10.1104/pp.105.070565. [Google Scholar] [CrossRef]
9. Jain, M., Tyagi, A. K., Khurana, J. P. (2008). Genome-wide identification, classification, evolutionary expansion and expression analyses of homeobox genes in rice. FEBS Journal, 275(11), 2845–2861. DOI 10.1111/j.1742-4658.2008.06424.x. [Google Scholar] [CrossRef]
10. Lai, W., Zhu, C., Hu, Z., Liu, S., Wu, H. et al. (2021). Identification and transcriptional analysis of zinc finger-homeodomain (ZF-HD) family genes in cucumber. Biochemical Genetics, 59(4), 884–901. DOI 10.1007/s10528-021-10036-z. [Google Scholar] [CrossRef]
11. Wang, H., Yin, X., Li, X., Wang, L., Zheng, Y. et al. (2014). Genome-wide identification, evolution and expression analysis of the grape (Vitis vinifera L.) zinc finger-homeodomain gene family. International Journal of Molecular Sciences, 15(4), 5730–5748. DOI 10.3390/ijms15045730. [Google Scholar] [CrossRef]
12. Niu, H., Xia, P., Hu, Y., Zhan, C., Li, Y. et al. (2021). Genome-wide identification of ZF-HD gene family in Triticum aestivum: Molecular evolution mechanism and function analysis. PLoS One, 16(9), e0256579. DOI 10.1371/journal.pone.0256579. [Google Scholar] [CrossRef]
13. Shi, Y., Pang, X., Liu, W., Wang, R., Su, D. et al. (2021). SlZHD17 is involved in the control of chlorophyll and carotenoid metabolism in tomato fruit. Horticulture Research, 8(1), 259. DOI 10.1038/s41438-021-00696-8. [Google Scholar] [CrossRef]
14. Wang, W., Wu, P., Li, Y., Hou, X. (2016). Genome-wide analysis and expression patterns of ZF-HD transcription factors under different developmental tissues and abiotic stresses in Chinese cabbage. Molecular Genetics and Genomics, 291(3), 1451–1464. DOI 10.1007/s00438-015-1136-1. [Google Scholar] [CrossRef]
15. Liu, H., Yang, Y., Zhang, L. (2021). Zinc Finger-Homeodomain transcriptional factors (ZF-HDs) in wheat (Triticum aestivum L.Identification, evolution, expression analysis and response to abiotic stresses. Plants, 10(3), 593. DOI 10.3390/plants10030593. [Google Scholar] [CrossRef]
16. Figueiredo, D. D., Barros, P. M., Cordeiro, A. M., Serra, T. S., Lourenço, T. et al. (2012). Seven zinc-finger transcription factors are novel regulators of the stress responsive gene OsDREB1B. Journal of Experimental Botany, 63(10), 3643–3656. DOI 10.1093/jxb/ers035. [Google Scholar] [CrossRef]
17. Xu, Y., Wang, Y., Long, Q., Huang, J., Wang, Y. et al. (2014). Overexpression of OsZHD1, a zinc finger homeodomain class homeobox transcription factor, induces abaxially curled and drooping leaf in rice. Planta, 239(4), 803–816. DOI 10.1007/s00425-013-2009-7. [Google Scholar] [CrossRef]
18. Mukherjee, K., Brocchieri, L., Burglin, T. R. (2009). A comprehensive classification and evolutionary analysis of plant homeobox genes. Molecular Biology and Evolution, 26(12), 2775–2794. DOI 10.1093/molbev/msp201. [Google Scholar] [CrossRef]
19. Zhang, X., Zong, J., Liu, J., Yin, J., Zhang, D. (2010). Genome-wide analysis of WOX gene family in rice, sorghum, maize, Arabidopsis and poplar. Journal of Integrative Plant Biology, 52(11), 1016–1026. DOI 10.1111/j.1744-7909.2010.00982.x. [Google Scholar] [CrossRef]
20. Zhao, Y., Zhou, Y., Jiang, H., Li, X., Gan, D. et al. (2011). Systematic analysis of sequences and expression patterns of drought-responsive members of the HD-Zip gene family in maize. PLoS One, 6(12), e28488. DOI 10.1371/journal.pone.0028488. [Google Scholar] [CrossRef]
21. Finn, R. D., Clements, J., Eddy, S. R. (2011). HMMER web server: Interactive sequence similarity searching. Nucleic Acids Research, 39(suppl_2), w29–w37. DOI 10.1093/nar/gkr367. [Google Scholar] [CrossRef]
22. Tvedebrink, T., Andersen, M. M., Curran, J. M. (2020). DNAtools: Tools for analysing forensic genetic DNA data. Journal of Open Source Software, 5(45), 1981. DOI 10.21105/joss.01981. [Google Scholar] [CrossRef]
23. Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M. et al. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 28(10), 2731–2739. DOI 10.1093/molbev/msr121. [Google Scholar] [CrossRef]
24. Wei, F., Coe, E., Nelson, W., Bharti, A. K., Engler, F. et al. (2007). Physical and genetic structure of the maize genome reflects its complex evolutionary history. PLoS Genetics, 3(7), e123. DOI 10.1371/journal.pgen.0030123. [Google Scholar] [CrossRef]
25. Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y. et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research, 30(1), 325–327. DOI 10.1093/nar/30.1.325. [Google Scholar] [CrossRef]
26. Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCt method. Methods, 25(4), 402–408. DOI 10.1006/meth.2001.1262. [Google Scholar] [CrossRef]
27. Khurana, P., Gaikwad, K. (2005). The map-based sequence of the rice genome. Nature, 436(7052), 793–800. DOI 10.1038/nature03895. [Google Scholar] [CrossRef]
28. Lin, Y. X., Jiang, H. Y., Chu, Z. X., Tang, X. L., Zhu, S. W. et al. (2011). Genome-wide identification, classification and analysis of heat shock transcription factor family in maize. BMC Genomics, 12(1), 1–14. DOI 10.1186/1471-2164-12-76. [Google Scholar] [CrossRef]
29. Kaul, S., Koo, H. L., Jenkins, J., Rizzo, M., Rooney, T. et al. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature, 408(6814), 796–815. DOI 10.1038/35048692. [Google Scholar] [CrossRef]
30. Wang, S., Chen, Y. (2019). Fine-tuning the expression of duplicate genes by translational regulation in Arabidopsis and maize. Frontiers in Plant Science, 10, 534. DOI 10.3389/fpls.2019.00534. [Google Scholar] [CrossRef]
31. Guo, H., Jiao, Y., Tan, X., Wang, X., Huang, X. et al. (2019). Gene duplication and genetic innovation in cereal genomes. Genome Research, 29(2), 261–269. DOI 10.1101/gr.237511.118. [Google Scholar] [CrossRef]
32. Li, L., Briskine, R., Schaefer, R., Schnable, P. S., Myers, C. L. et al. (2016). Co-expression network analysis of duplicate genes in maize (Zea mays L.) reveals no subgenome bias. BMC Genomics, 17(1), 875. DOI 10.1186/s12864-016-3194-0. [Google Scholar] [CrossRef]
33. Liu, M., Wang, X., Sun, W., Ma, Z., Zheng, T. et al. (2019). Genome-wide investigation of the ZF-HD gene family in Tartary buckwheat (Fagopyrum tataricum). BMC Plant Biology, 19(1), 248. DOI 10.1186/s12870-019-1834-7. [Google Scholar] [CrossRef]
34. Bataillon, T., Gauthier, P., Villesen, P., Santoni, S., Thompson, J. D. et al. (2022). From genotype to phenotype: Genetic redundancy and the maintenance of an adaptive polymorphism in the context of high gene flow. Evolution Letters, 6(2), 189–202. DOI 10.1002/evl3.277. [Google Scholar] [CrossRef]
35. Peng, J. (2019). Gene redundancy and gene compensation: An updated view. Journal of Genetics and Genomics, 46(7), 329–333. DOI 10.1016/j.jgg.2019.07.001. [Google Scholar] [CrossRef]
36. Nieto Feliner, G., Casacuberta, J., Wendel, J. F. (2020). Genomics of evolutionary novelty in hybrids and polyploids. Frontiers in Genetics, 11, 792. DOI 10.3389/fgene.2020.00792. [Google Scholar] [CrossRef]
37. Kuzmin, E., Taylor, J. S., Boone, C. (2022). Retention of duplicated genes in evolution. Trends in Genetics, 38(1), 59–72. DOI 10.1016/j.tig.2021.06.016. [Google Scholar] [CrossRef]
38. Bollier, N., Gonzalez, N., Chevalier, C., Hernould, M. (2022). Zinc finger-homeodomain (ZHD) and mini zinc finger (MIF) proteins are key players in plant growth and responses to environmental stresses. Journal of Experimental Botany, 73, erac194. DOI 10.1093/jxb/erac194. [Google Scholar] [CrossRef]
39. Zhou, C., Zhu, C., Xie, S., Weng, J., Lin, Y. et al. (2021). Genome-wide analysis of zinc finger motif-associated homeodomain (ZF-HD) family genes and their expression profiles under abiotic stresses and phytohormones stimuli in tea plants (Camellia sinensis). Scientia Horticulturae, 281(2), 109976. DOI 10.1016/j.scienta.2021.109976. [Google Scholar] [CrossRef]
40. Jain, M., Khurana, P., Tyagi, A. K., Khurana, J. P. (2008). Genome-wide analysis of intronless genes in rice and Arabidopsis. Functional & Integrative Genomics, 8(1), 69–78. DOI 10.1007/s10142-007-0052-9. [Google Scholar] [CrossRef]
41. Yan, H., Zhang, W., Lin, Y., Dong, Q., Peng, X. et al. (2014). Different evolutionary patterns among intronless genes in maize genome. Biochemical and Biophysical Research Communications, 449(1), 146–150. DOI 10.1016/j.bbrc.2014.05.008. [Google Scholar] [CrossRef]
42. Bänziger, M., Araus, J. L. (2007). Recent advances in breeding maize for drought and salinity stress tolerance. Advances in Molecular Breeding toward Drought and Salt Tolerant Crops, 587–601. DOI 10.1007/978-1-4020-5578-2_23. [Google Scholar] [CrossRef]
43. Barth, O., Vogt, S., Uhlemann, R., Zschiesche, W., Humbeck, K. (2009). Stress induced and nuclear localized HIPP26 from Arabidopsis thaliana interacts via its heavy metal associated domain with the drought stress related zinc finger transcription factor ATHB29. Plant Molecular Biology, 69(1–2), 213–226. DOI 10.1007/s11103-008-9419-0. [Google Scholar] [CrossRef]
44. Nakashima, K., Yamaguchi-Shinozaki, K. (2006). Regulons involved in osmotic stress-responsive and cold stress-responsive gene expression in plants. Physiologia Plantarum, 126(1), 62–71. DOI 10.1111/j.1399-3054.2005.00592.x. [Google Scholar] [CrossRef]
45. Zhao, T., Wang, Z., Bao, Y., Zhang, X., Yang, H. et al. (2019). Downregulation of SL-ZH13 transcription factor gene expression decreases drought tolerance of tomato. Journal of Integrative Agriculture, 18(7), 1579–1586. DOI 10.1016/S2095-3119(19)62621-3. [Google Scholar] [CrossRef]
46. Tran, L. S. P., Nakashima, K., Fujita, Y., Sakuma, Y., Maruyama, K. et al. (2003). Functional analysis of the members of Arabidopsis NAC transcription factors controlling the expression of the ERD1 gene under drought stress. Plant and Cell Physiology Supplement, 44, 74. DOI 10.14841/jspp.2003.0.74.0. [Google Scholar] [CrossRef]
Appendix A
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools