 Open Access
Open Access
ARTICLE
A Novel Dominant Allele from 93-11, ES(4), Represses Reactive Oxygen Species Scavenging and Leads to Early-Senescence in Rice
Key Laboratory of Crop Physiology, Ecology and Genetic Breeding, Ministry of Education, Jiangxi Agricultural University, Nanchang, 330045, China
* Corresponding Author: Jie Xu. Email:
(This article belongs to the Special Issue: High-Yield Rice Physiology & Genetics)
Phyton-International Journal of Experimental Botany 2023, 92(3), 665-677. https://doi.org/10.32604/phyton.2023.025266
Received 02 July 2022; Accepted 25 August 2022; Issue published 29 November 2022
Abstract
Senescence is the last developmental process in plant, which has an important impact on crop yield and quality. In this study, a stable hereditary early-senescence line BC64 was isolated from the high-generation recombinant inbred lines of 93-11 and Wuyunjing7 (W7). Genetic analysis showed that the premature aging phenotype was controlled by a dominant gene derived from 93-11. By linkage analysis, the gene was primarily mapped in the region between marker B4 and B5 near the centromere of chromosome 4, described as ES(4). Through multiple backcrossing with W7, the near-isogenic line of ES(4) (NIL-ES(4)) was obtained. Compared with wild-type W7, NIL-ES(4) plants showed more sever senescence phenotype in both nature and dark conditions. In NIL plants, leaves turned yellow at the fully tillering stage; photosynthetic rate, pollen fertility and seed setting rate were decreased. Moreover, the malondialdehyde, proline content and relative conductivity in NIL-ES(4) were significantly higher than those in W7; both transcript level and activities of reactive oxygen species scavenging enzymes were repressed; H2O2 and O2− were significantly accumulated. This study provides a basis for further cloning and function identification of ES(4).Keywords
Rice is one of the most important food crops, and half population of the world take it as the staple food. The growth of rice yield has further slowed down during the first decade of this century. The rice-consuming countries population continues to increase rapidly at a rate of over 1.5% per year, leading to a growing demand for rice. Therefore, improvement of grain yield and grain quality are of key agricultural importance [1]. Senescence is the final step of plant growth and development. Initiation timing of senescence has great influences on the plant biomass and quality formation. In crop plants, early senescence strongly affects grain filling, directly limit grain yield and quality [2,3]. Therefore, defining the molecular and physiological mechanisms of early senescence in rice are of great significance to enhance grain yield and quality.
Leaf senescence is a very complicated process, a range of internal and external factors appear to cause leaf senescence [4–6]. The process of leaf senescence is accompanied by a series of changes in cell structure and physiology of plants, including changes in chlorophyll, protein, photosynthetic efficiency, reactive oxygen species (ROS) scavenging and accumulation, malondialdehyde (MDA), proline and other indicators [7–9]. Thus, a large number of genes play essential roles in leaf senescence, and many senescence-associated genes have been identified in rice. These genes are found to function in several different biological processes, such as chlorophyll synthesis and degradation, chloroplast development, ROS scavenging system, programmed cell death and biotic and abiotic stress responses [10–13]. Chlorophyll is an essential molecule for harvesting light energy during photosynthesis. Therefore, genes involved in either chlorophyll biosynthesis or degradation are responsive to leaf senescence. The mutants of OsPORB [11,14], OsCAO1 [10], OsNOL, OsNYC1 [15,16], OsNYC3 [17], OsNYC4 [18], OsPAO and OsRCCR1 [19] have been reported to have early-senescence phenotypes, as well as low grain yield and quality. In additon, many genes involved in the chlorophyll metabolism or other important processes in chloroplast development, also could induce senescence, such as OsPSL [12], OsARVL4 [20], OsNDPK2 [21], OsCRTISO [22], WFSL1 [23], Heme Oxygenase 1 [24], ES1 [25] and so on. ROS scavenging is repressed and ROS is accumulated in all these mutants. Moreover, most early-senescence mutants are more sensitive to biotic and abiotic stresses [10,21,24,25]. Although many senescence associative genes have been identified, more genes and mutants are still necessary to further understand molecular mechanism of senescence because this process is truly complicated.
In this study, BC64 with stable early senescence phenotype, was isolated from the high-generation RILs of 93-11 and wuyunjing 7 (W7). This line appeared severe premature before heading. Genetic analysis revealed that the early senescence phenotype was controlled by a dominant gene which was derived from 93-11; and the gene was mapped to the near-centromere region on chromosome 4, named ES(4). We performed phenotypic identification, physiological and biochemical analysis to explore its function and mechanism in regulating leaf senescence.
2.1 Plant Materials and Growth Conditions
ES(4) is a stable hereditary line, BC64, with early senescence phenotype, which was found in the high-generation RILs of 93-11 and W7. The near isogenic lines for ES(4) (NIL-ES(4)) were developed from the BC4F2 generation by repetitive backcrossing with W7. All plants were grown in the paddy field in Jiangxi Agricultural University, Nanchang, Jiangxi Province, China.
For dark-induced senescence assay, the flag leaves were harvested from the NIL and W7 plants, and were cut into small size, about 1 cm. Then the samples were incubated in MES buffer (30 mM, pH 5.8) and kept in darkness at 28°C for 4 days.
2.2 Genetic Analysis and Linkage Analysis
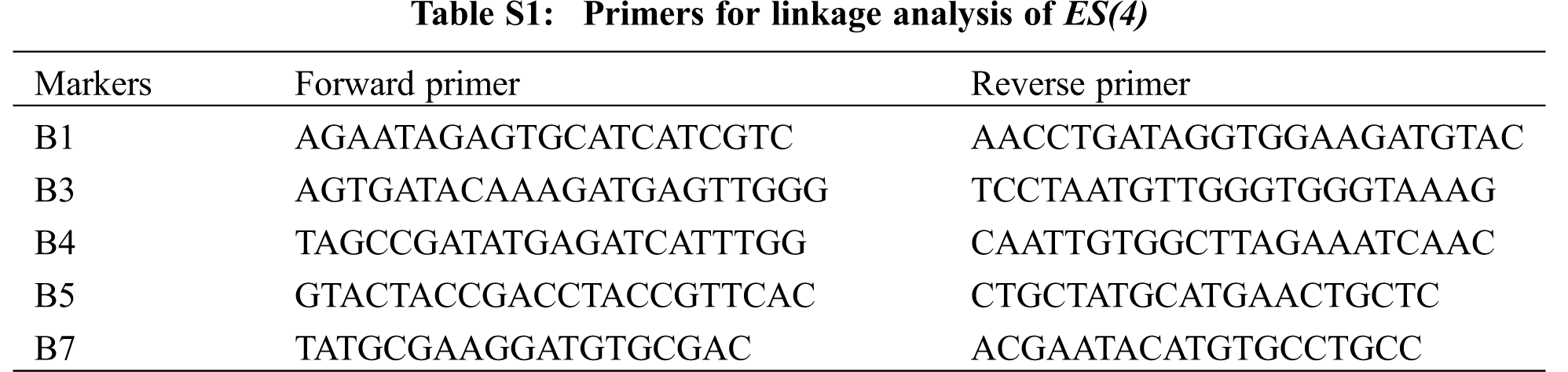
For genetic analysis, BC64 was crossed with its parents lines respectively, and the phenotype of F1 and character separation ratio of F2 were detected. The F2 population of BC64 and W7 were used for linkage analysis. The DNA extraction of the mixed pool for genetic analysis and the F2 individuals of gene mapping were performed by CTAB method. For linkage analysis, a DNA bulk poor from 23 F2 individuals with normal leaves was screened with a total of 265 simple sequence repeat (SSR) markers and sequence tagged site (STS) markers uniformly distributed on all 12 chromosomes in our laboratory. Subsequently, 274 individuals were used for confirmation of the linkage and genetic distance estimation. The PCR products were seperated by 8%–12% polyacrylamide gel and 4%–5% agarose gel, and the band results were stained by silver nitrate and gelred, respectively. The molecular markers were listed in Table S1.
2.3 Chlorophyll Content Analysis
Chlorophyll content of W7 and NIL-ES(4) flag leaves were examined at the fully tillering stage. The leaves were cut into pieces of about 1 cm, and 1 g of leaves were soaked in 80% acetone in darkness for 12 h with gentle shake. The extract was collected for colorimetric determination. The total Chl were estimated with spectrophotometric values of 470, 645 and 663 nm.
2.4 Quantification of Senescence-Related Physiological Indicators
The flag leaves of W7 and NIL-ES(4) were selected for the determination of their physiological indexes at the fully tillering stage. Peroxidase (POD) activity, superoxide dismutase (SOD) activity, catalase (CAT) activity, MDA content, and proline content, were measured by using corresponding kits of Jiangsu KeMing Biological Company (POD-1-Y, SOD-1-Y, CAT-1-Y, MDA-1-Y, and PRO-1-Y, respectively). The specific method is described in the manual, and the measure of each indicator was repeated three times. Conductivity was determined as previously described [10]. The net photosynthetic rate was measured by using a CI-340 portable photosynthetic apparatus (CID, USA) in the presence of sufficient sunlight at the filling stages.
2.5 Hydrogen Peroxide and Superoxide Anion Detection
The accumulation of H2O2 in leaves was detected by diaminobenzidine (DAB) staining (0.5 mg/mL in 10 mM potassium phosphate buffer, pH 7.6); NBT (1 mg/ml in 50 mM Tris acetate buffer, pH 5.8) was applied to detect O2−. The fresh leaves of W7 and NIL-ES(4) were harvested separately, then the leaves were immersed into the staining buffer. After staying in darkness for 8 h, the leaves were transferred to alcohol and boiled to remove chlorophyll.
2.6 RNA Extraction and Quantitative Real-Time PCR
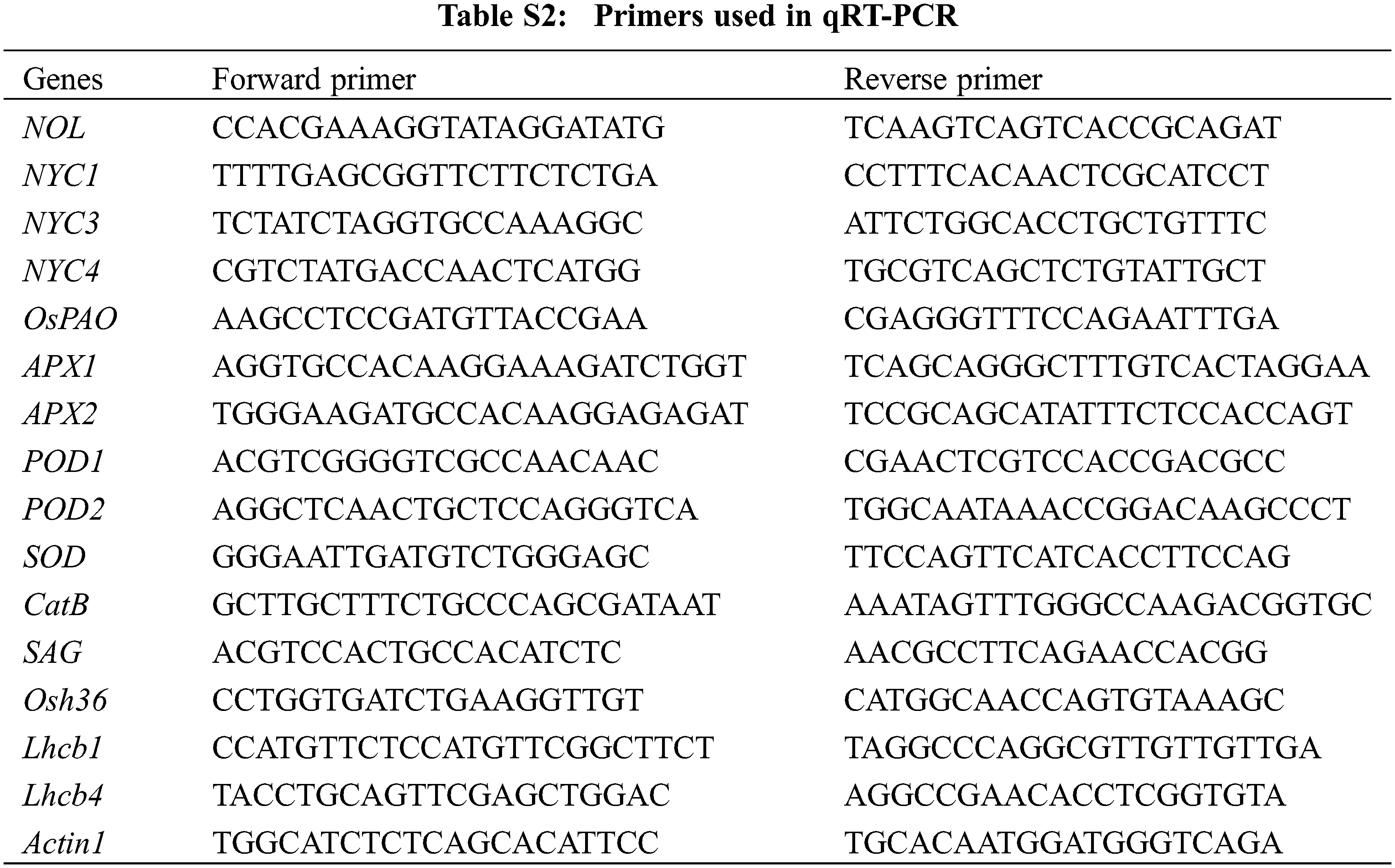
The leaves of W7 and NIL-ES(4) were collected at the fully tillering stage or filling stage. Total RNA extraction was performed using a TaKaRa MiniBEST Plant RNA Extraction Kit (TAKARA, Code No. 9769). All the samples were digested by DNase I (TAKARA, Code No. 2270A). Reverse transcription was performed with PrimeScript™ II 1st Strand cDNA Synthesis Kit (Code No. 6210A). 2 × SYBR Green PCR Master Mix (Applied Biosystems) was applied for real-time PCR, the reaction was carried out in a two-step process using a PCR procedure of 95°C for 1 min, 95°C for 15 s, 60°C for 30 s, and 40 cycles (ABI 7900). The transcription level of these genes (including OsLchb1, OsLchb4, OsNOL, OsNYC1, OsNYC3, OsNYC4, OsPAO, OsRCCR1, OsAPX1, OsAPX2, OsPOD1, OsPOD2, OsCatB, OsSAG, Osh36 and Actin1) were quantified, The primers for qRT-PCR were listed in Table S2.
RNA or protein samples used for detection of chlorophyll content, electrolyte leakage, senescence-related physiological indicators and expression levels of related genes were obtained from bulk samples of three independent plants. The 2−ΔCt and 2−ΔΔCt methods were used for gene expression analysis. All of the experiments were performed in triplicate, the error bars indicate the standard error of the mean. For statistical analysis of photosynthetic rate, mean and error bars from at least 10 plants are indicated on the y axes.
3.1 Genetic Analysis and Linkage Analysis of the Early-Senescence Gene
We obtained an early-senescence line (BC64) from the progeny of the high-generation RILs of 93-11 and W7. The BC64 showed an obvious early-senescence phenotype at full tillering, leaves turned yellow, and the height was significantly lower and tiller number was less than its parents (Fig. 1A). In order to elucidate the genetic pattern of the early-senescence phenotype of BC64, this line was crossed with each parent, respectively. The F1 hybrid plants of both crossings showed a premature aging phenotype (F1 of W7 and BC64 was shown in Fig. 1B, F1 of 93-11 and BC64 was not shown). All F2 individuals of 93-11 and BC64 showed different degrees of premature aging phenotype (data not shown), while the F2 population generated from crossing W7 with BC64 showed separation in the target trait. We obtained 274 normal individuals from a total of 1132 F2 plants from crossing of BC64 with W7. The ratio of individuals with a normal phenotype to those with an ES phenotype showed a good fit to the expected value [i.e., 1:3 (χ2 = 0.382, P = 0.537)]. All these results indicated that the early-senescence phenotype was controlled by a single dominant gene derived from 93-11. We found that the tip of 93-11 leaves also appeared yellow before heading, showing a certain senescence trait (Fig. 1A). Moreover, the hybrid rice varieties generated from 93-11 as both male and female parents, have a significant senescence at the late stage, indicating that there was a dominant premature senescence gene on the 93-11 genome, which was completely consistent with our results in this study.
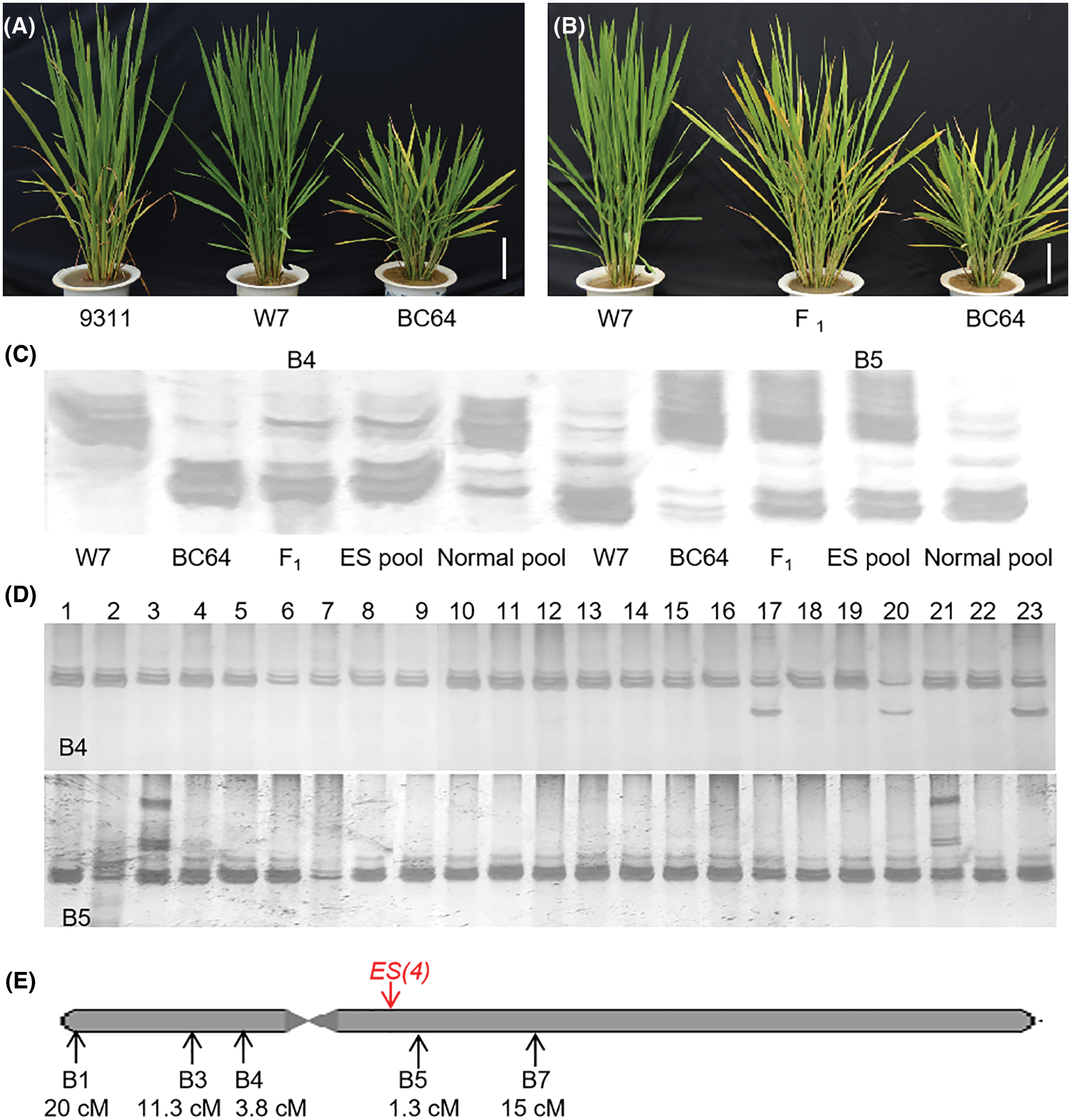
Figure 1: Primary mapping of early senescence gene ES(4). (A) Morphological comparison of BC64 and its parents at the tillering stage; (B) phenotype of F1 plants of BC64 and W7; (C) ES(4) was linked to the molecular markers B4 and B5; (D) genotype analysis of F2 individuals with normal phenotype; (E) the genetic distance analysis of linked markers and ES(4). Scale bar = 10 cm
274 normal plants from the same F2 population were used for linkage analysis and genetic distance estimation. Linkage analysis was carried out by the BSA method. The results showed that the early-senescence phenotype was linked to the molecular markers B3, B4 and B5 (Fig. 1C). The genotype analysis of F2 individuals confirmed the linkage, and indicated that ES(4) was located in the interval between B4 and B5 which contains the centromere (Fig. 1D). The genetic distances between ES(4) and molecular markers were calculated with recombinant gamete ratio. There were 110, 62, 21, 7, 82 recombinant gametes in the total 548 gametes at the B1, B3, B4, B5, B7 sites, indicating that the genetic distances between ES(4) and them were 20, 11.3, 3.8, 1.3 and 15 cm, respectively (Fig. 1E).
3.2 Development of Near-Isogenic Line NIL-ES(4)
According to the complicated genetic background of RILs, eliminating the interference of different varieties is crucial to identify the function of ES(4). Therefore, the near isogenic line NIL-ES(4) in W7 background was developed by continuously backcrossing. Compared with W7, the height was lower and tiller number was less in NIL-ES(4) (Fig. 2A). NIL-ES(4) plants showed severe premature aging before heading, the leaf turned yellow rapidly (Fig. 2A), the chlorophyll content was much lower in NIL plants (Fig. 2D). The electric conductivity of the blade was increased significantly in NIL-ES(4), increased by about 60% of that in W7 (Fig. 2E), indicating that the NIL-ES(4) cells were damaged seriously, leading to early senescence. The photosynthesis rate of leaves at the filling stage in the NIL plants was severely inhibited, it was only about one forth of that in W7, which might be caused by chlorophyll degradation and senescence (Fig. 2F). The expression levels of OsLhcb1 and OsLhcb4, two important photosynthesis-associated genes, were also examined. The results showed that OsLhcb1 and OsLhcb4 were strongly down-regulated in NIL-ES(4) compared with W7 (Figs. 2E and 2F). Additionally, the pollen fertility of NIL-ES(4) was partially reduced; about 14.7% pollen was abortive, resulting in a low seed setting rate (Figs. 2B and 2C).
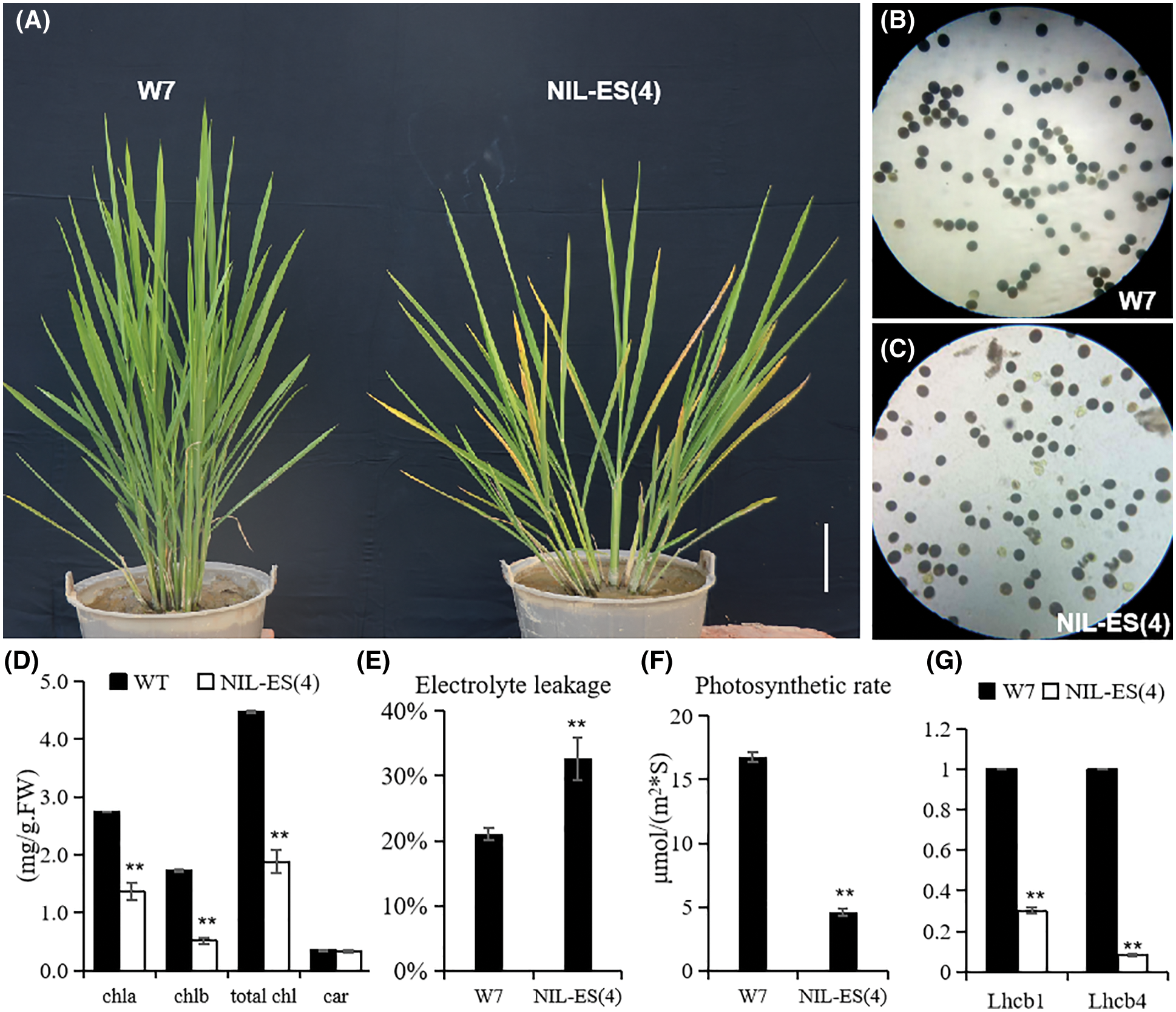
Figure 2: Construction and phenotype identification of NIL-ES(4). (A) The senescence phenotype of NIL-ES(4); (B and C) pollen fertility comparison between W7 and NIL-ES(4); (D, E) chlorophyll content (D) and electrolyte leakage (E) in W7 and NIL-ES(4) at the tillering stage; (F and G) photosynthetic rate (F) and expression level photosynthesis-associated gene (G) in W7 and NIL-ES(4) flag leaves at the filling stage. Error bars indicate SD; n = 20 plants. * and ** represented a significant difference (P < 0.05) and an extremely significant difference (P < 0.01), respectively
3.3 Relative Expression of Senescence-Associated Genes
In order to further explore the functions of ES(4) in senescence, we detected the expression level of senescence-associated genes, including chlorophyll degradation-related genes, ROS scavenging-associated genes and senescence marker genes. Among five chlorophyll degradation-related genes, OsNYC3 and OsPAO were significantly up-regulated in NIL-ES(4) compared with W7 (Figs. 3C and 3E), while OsNOL, OsNYC1 and OsNYC4 were down-regulated (Figs. 3A, 3B and 3D). Thus, ES(4) might mainly participate in the chlorophyll degradation pathway by regulating OsNYC3 and OsPAO; the chlorophyll content was much lower in NIL plants, suggested that even smaller amount of these genes were enough for chlorophyll degradation. Therefore, ES(4) might not directly affect chlorophyll degradation to induce premature.
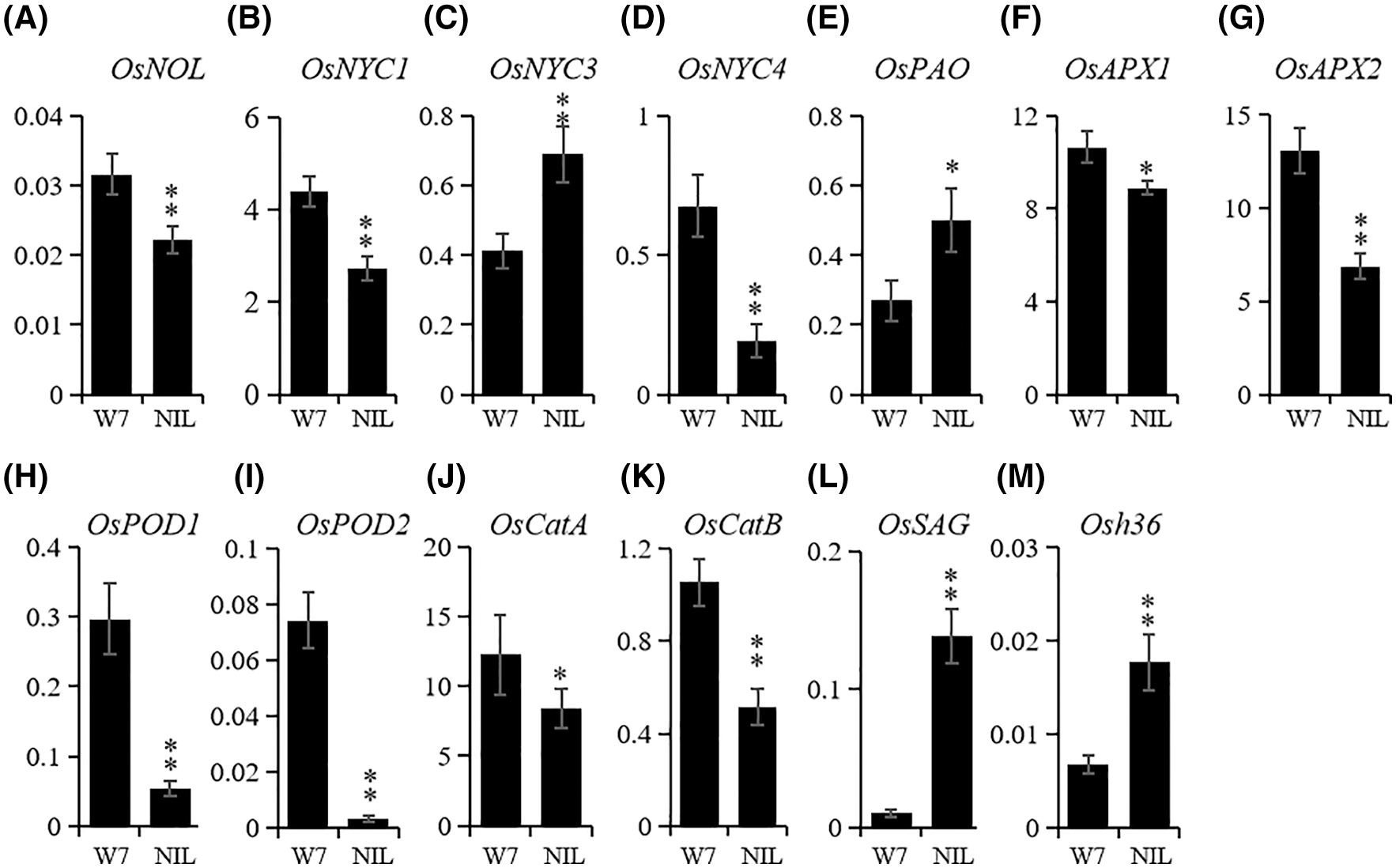
Figure 3: The expression differences of senescence-associated genes between W7 and NIL-ES(4). (A–E), transcription level of chlorophyll degradation related genes: OsNOL (A), OsNYC1 (B), OsNYC3 (C), OsNYC4 (D) and OsPAO (E); (F–K), transcription level of ROS scavenging related genes: OsAPX1 (F), OsAPX2 (J), OsPOD1 (H), OsPOD2 (I), OsCatA (J), and OsCatB (K); (L and M) expression of senescence-induced genes, OsSAG (K) and Osh36 (I). Actin1 was used as a reference gene. The mean of each expression was based on the average of three biological repeats calculated using the relative quantification method; leaves from more than 10 plants were used for qRT-PCR to avoid individual difference. * and ** represented the significant difference (P < 0.05) and extremely significant difference (P < 0.01), respectively
All ROS-scavenging-related genes which were detected in this study, including OsAPX1, OsAPX2, OsPOD1, OsPOD2, OsCatA and OsCatB, were down-regulated in NIL-ES(4), especially OsPOD1 and OsPOD2 (Figs. 3F–3K). The expression levels of OsPOD1 and OsPOD2 in NIL-Es(4) were less than 1/4 and 1/32 of those in W7, respectively (Figs. 4H and 4I). Two typical cell senescence marker genes, OsSAG and Osh36, whose expression were senescence-induced [26,27]; and their transcriptional levels were obviously increased in NIL plants. The OsSAG was sharply up-regulated, 32-fold of that in W7, indicating the serious senescence in NIL-ES(4) (Figs. 3L and 3M). These results suggested that ES(4) repressed the ROS scavenging associated genes and then caused early senescence.
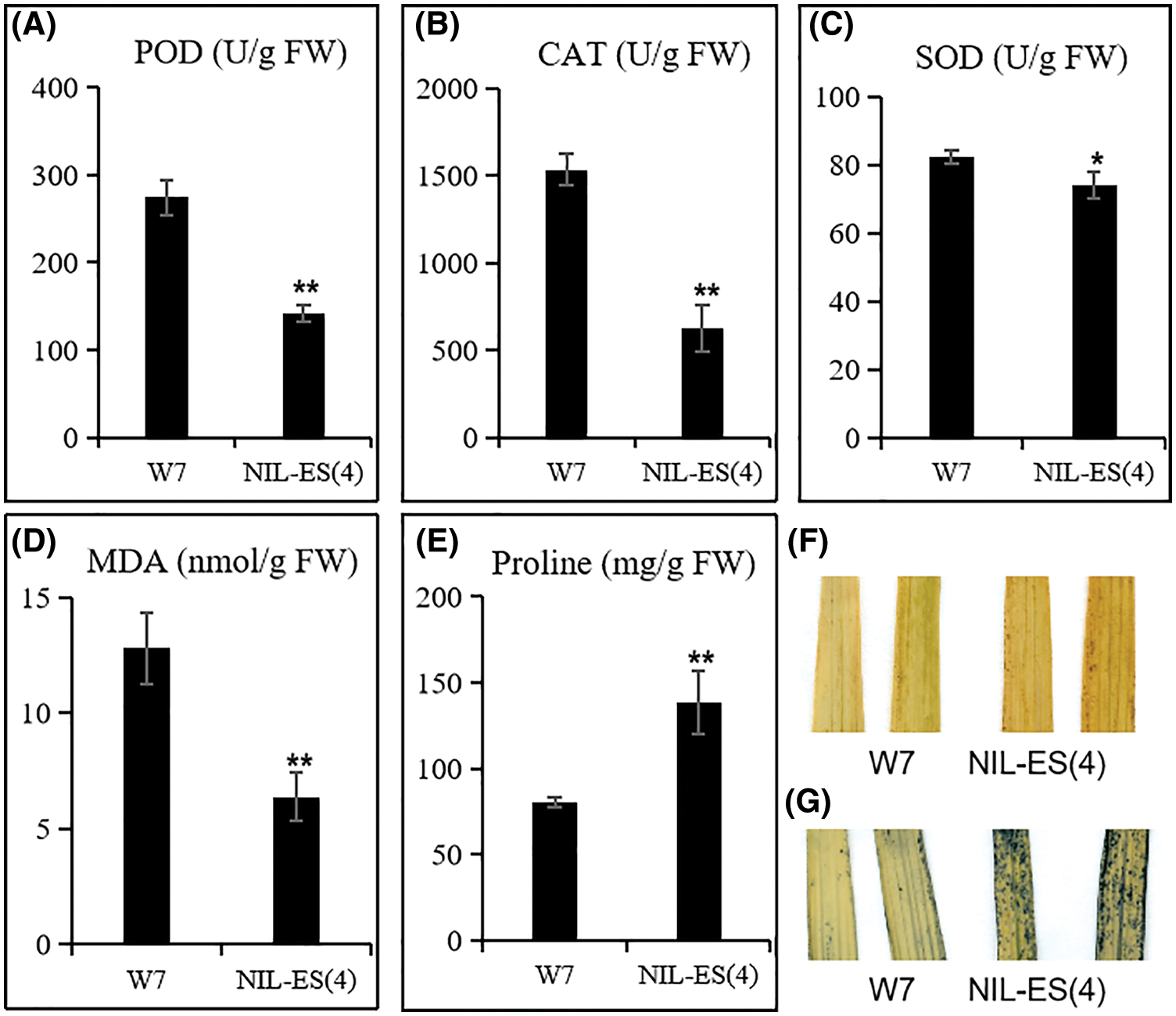
Figure 4: Determination of physiological characteristics of senescence. POD activity (A), CAT activity (B), SOD activity (C), MDA content (D) and Proline content (E) were quantified in W7 and NIL-ES(4); H2O2 (F) and O2− (G) were detected with DAB and NBT. Error bars indicate SD; n = 20 plants. * and ** represented a significant difference (P < 0.05) and an extremely significant difference (P < 0.01), respectively
3.4 Determination of Physiological Characteristics of Senescence
Several indexes of leaf senescence were detected at the tillering stage. The enzymatic activities of SOD, CAT and POD in NIL-ES(4) leaves were significantly lower than those in W7 leaves; the activities of CAT and POD were decreased by 59% and 39%, respectively (Figs. 4A–4C). These results revealed that the ROS scavenging ability in NIL-ES(4) leaves was seriously decreased, which is consistent with the down-regulated expression levels of ROS scavenging related genes (Figs. 3F–3K). Subsequently, we tested the accumulation of H2O2 and O2− (Figs. 4F and 4G) in leaves with NBT and DAB staining methods. More dark spots/tissues appeared after either DAB or NBT staining, indicating that more H2O2 and O2− were accumulated in NIL-ES(4). In addition, MDA and proline were quantified. Compared with W7, the content of MDA was increased by 52%, and proline content was increased by 72% in NIL plants. These results suggested that the leaves of NIL plants showed more severe cell damage. Taken together, ES(4) affected the expression of ROS scavenging-related gene, then the activity of the ROS scavenging enzymes was decreased, resulting in the accumulation of ROS, cell death, and finally accelerated senescence.
3.5 ES(4) Accelerates Dark-Mediated Senescence
Besides premature under natural conditions, dark-induced senescence was also examined. The detached leaves from the NIL and W7 plants were incubated in MES buffer and kept under darkness conditions at 28°C. Chlorophyll was quantified at 0, 2 and 4 days after the dark treatment separately. Although the initial chlorophyll content was lower in NIL-ES(4) leaves (1.87 mg/g·FW in NIL and 4.46 mg/g·FW in W7), the chlorophyll degradation rate was faster in NIL-ES(4) than that in W7, especially after 2 days (Figs. 5A–5C). After 4 days, the leaves from NIL turned almost completely yellow; the chlorophyll content was as low as 0.67 mg/g·FW, while 2.79 mg/g·FW in W7; Compared with the initial contents, only 35.8% chlorophyll was left in NIL-ES(4) leaf pieces, while 62.6% in W7 leaves. These results indicated that ES(4) in NIL plants could also accelerate dark-mediated senescence.

Figure 5: The chlorophyll degradation in dark induced senescence. Chlorophyll a (A), Chlorophyll b (B) and total chlorophyll content (C) at 0, 2 and 4 d after dark treatment. Error bars indicate SD; n = 5 plants. * and ** represented a significant difference (P < 0.05) and an extremely significant difference (P < 0.01), respectively
Early senescence is one of the most important factors which affect rice yield and quality [10,28]. Senescence in rice was reviewed by Leng et al. [29], many mutants were identified such as es1 [25], pgl [10], and psl3 [30]. Most of the identified mutants are controlled by recessive genes, while the dominant gene-controlled early senescence mutants (e.g., psl3) are very rare. On the other hand, Indica cv. 93-11 is a popular variety widely grown in China. It has many advantages: stem stout, erect leaves, perfect plant type, good grain quality, stable yield and lodging resistance. Moreover, 93-11 is the paternal cultivar of a super-hybrid rice, Liang-You-Pei-Jiu (LYP9), which has 20% to 30% more yield per hectare than many ordinary rice varieties. However, 93-11 has a light early senescence phenotype after heading (Fig. 1A), and hybrid rice varieties from crossing 93-11 with other varieties also have the premature phenomenon, indicating that there is a dominant gene causing early senescence in 93-11. Thus, cloning and functional identification of this senescence-related gene is beneficial for rice breeding.
It is well known that crossingover rarely happen near the centromere. Therefore, it is truly hard to narrow the region containing the target gene by map-based cloning method, leading to a related large interval in our experiments (Fig. 1E). However, only a few genes have been cloned and their functions were identified; among them, several genes encode definite or potential senescence-related factors. OsSRT1 encodes a NAD+-dependent histone deacetylase, and negatively regulates leaf senescence by repressing expression of downstream genes such as OsPME1 [31]; its knockdown plants increase the acetylation and decrease the dimethylation of H3K9, resulting in H2O2 production, DNA fragmentation and cell death [31,32]. OsAPX3 encodes an ascorbate peroxidase which catalyses the conversion of H2O2 to H2O and O2 using ascorbate as the specific electron donor [33]. APXs play essential roles in scavenging and in protecting cells against the toxic effects of H2O2 in higher plants [34]. OsGRX12 is a member of glutaredoxins which are ubiquitous oxidoreductase enzymes; they mediate the reduction of disulphide bonds of substrates in presence of glutathione. These enzymes are involved in diverse cellular processes and play an important role in defense against oxidative stress [35].
According to genetic analysis, ES(4) was proved to be a novel dominant gene inducing senescence in BC64 (Fig. 1A and 1B). Most of the identified senescence mutants are controlled by recessive genes, the dominant gene-controlled early senescence mutants are very rare. PSL3 was also a dominant gene, this gene was mapped to an interval of 53.5 kb on chromosome 7. In psl3, the chloroplast number of mesophyll cells decreased, some chloroplasts membranes were dissolved and the thylakoid was disorderly arranged; the SOD activities were significantly decreased in the senescence part of the psl3 mutant. However, the PSL3 gene has not been already cloned, so the regulatory mechanism of PSL3 in leaf senescence is not clear [30].
Over 185 senescence-associated genes have been identified in rice [30]; they are mainly involved in chloroplast development and degradation [10,36], phytohormones signal transduction [37,38], and energy metabolism [32,39], and nitrogen remobilization [40,41]. During leaf senescence, chloroplast degradation usually induces the production of ROS. On the other hand, hyperaccumulation of ROS in plant cells cause severe damage to lipids, proteins, and DNA [42,43], which would induce chloroplast degradation. Therefore, production and scavenging of ROS must be tightly controlled by the antioxidant defense system to maintain a dynamic balance of the ROS level in plants [44,45]. ROS (O2− and H2O2) was accumulated in NIL-ES(4) leaves, which would result in severe damage to the proteins, membrane lipids, DNA and other cellular components of plants [46]. Leaves turned yellow earlier and chlorophyll content was lower in NIL-ES(4) than in WT plants. We detected five genes involved in chlorophyll degradation; only OsNYC3 and OsPAO were up-regulated in NIL-ES(4) (Figs. 3C and 3E), and the rest were down-regulated (Figs. 3A, 3B and 3D). Nevertheless, both transcriptional levels (Figs. 3F–3K) and enzymatic activity (Figs. 4A–4C) of the major antioxidant enzymes (including CAT, SOD, POD, APX) were significantly decreased in NIL-ES(4). Taken together, ES(4) are much more likely to directly influence ROS scavenging than production (e.g., chloroplast degradation).
Chlorophyll is a key component in the plant photosynthesis system; the transcription level of the photosynthesis-associated genes (OsLhcb1 and OsLhcb4) and the photosynthesis rate were reduced which might result from the yellowing of leaves in NIL plants (Figs. 2E–2G); MDA is a substance produced by the oxidation of cell membrane lipids which caused by the accumulate of reactive. MDA is the final product of membrane lipid peroxidation which caused the accumulation of ROS, and proline has been reported to be a stabilizer to protect cells [47,48], so MDA and proline are direct signs of cell death. Accumulation of both MDA and proline together with increased expression of OsSAG and Osh36 (senescence-induced genes) indicated more severe cell damage in NIL-ES(4) plants (Figs. 4K, 4M, 4D and 4E). Finally, plants are more sensitive to internal or external disadvantageous factors at the reproductive stage, especially pollen development. So, most early senescence mutants have low fertility [22,25,49]. Both BC64 and NIL-ES(4) had very low fertility (Fig. 2C), which might be caused by ROS accumulation, leading to low seed setting rate.
The widely grown Indica cv. 93-11 harbour a dominant senescence-related gene, ES(4). ES(4) is located near the centromere of chromosome 4. The ES(4) allele in 93-11 repress the expression of ROS scavenging related genes, and increase ROS accumulation, leading to cell death and early senescence. However, further studies are required to clone ES(4) gene and investigate its molecular mechanism in regulating senescence.
Authorship: Jie Xu conceived the idea and supervised the work. Zhishu Jiang, Cong Gan, Yulian Liu, Xiaoli Lin, Limei Peng, Yongping Song and Xiaowei Luo performed experiments and analysed data, and Zhishu Jiang and Jie Xu wrote and revised the manuscript.
Funding Statement: This research was supported by grants from the Science Foundation of Jiangxi Province (Grant No. 20212ACB215003) and the National Natural Science Foundation of China (Grant No. 31960403).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Khush, G. S. (2005). What it will take to feed 5.0 billion rice consumers in 2030. Plant Molecular Biology, 59(1), 1–6. [Google Scholar]
2. Yoo, S. C., Cho, S. H., Zhang, H., Paik, H. C., Lee, C. H. et al. (2007). Quantitative trait loci associated with functional stay-green SNU-SG1 in rice. Molecules and Cells, 24(1), 83–94. [Google Scholar]
3. Gregersen, P. L., Culetic, A., Boschian, L., Krupinska, K. (2013). Plant senescence and crop productivity. Plant Molecular Biology, 82(6), 603–622. [Google Scholar]
4. Gully, K., Hander, T., Boller, T., Bartels, S. (2015). Perception of Arabidopsis AtPep peptides, but not bacterial elicitors, accelerates starvation-induced senescence. Frontiers in Plant Science, 6, 14. [Google Scholar]
5. Feller, U. (2016). Drought stress and carbon assimilation in a warming climate: Reversible and irreversible impacts. Journal of Plant Physiology, 203, 84–94. [Google Scholar]
6. Liebsch, D., Keech, O. (2016). Dark-induced leaf senescence: New insights into a complex light-dependent regulatory pathway. New Phytologist, 212(3), 563–570. [Google Scholar]
7. Maillard, A., Diquélou, S., Billard, V., Laîné, P., Garnica, M. et al. (2015). Leaf mineral nutrient remobilization during leaf senescence and modulation by nutrient deficiency. Frontiers in Plant Science, 6, 317. [Google Scholar]
8. Krautler, B. (2016). Breakdown of chlorophyll in higher plants-phyllobilins as abundant, yet hardly visible signs of ripening, senescence, and cell death. Angewandte Chemie International Edition, 55(16), 4882–4907. [Google Scholar]
9. Thakur, N., Sharma, V., Kishore, K. (2016). Leaf senescence: An overview. Indian Journal of Plant Physiology, 21, 225–238. [Google Scholar]
10. Yang, Y. L., Xu, J., Huang, L. C., Leng, Y. J., Dai, L. P. et al. (2016). PGL, encoding chlorophyllide a oxygenase 1, impacts leaf senescence and indirectly affects grain yield and quality in rice. Journal of Experimental Botany, 67(5), 1297–1310. [Google Scholar]
11. Yang, Y. L., Xu, J., Rao, Y. C., Zeng, Y. J., Liu, H. J. et al. (2016). Cloning and functional analysis of pale-green leaf (PGL10) in rice (Oryza sativa L.). Plant Growth Regulation, 78, 69–77. [Google Scholar]
12. Ke, S., Liu, S., Luan, X., Xie, X. M., Hsieh, T. F. et al. (2019). Mutation in a putative glycosyltransferase-like gene causes programmed cell death and early leaf senescence in rice. Rice, 12(1), 1–14. [Google Scholar]
13. Tamary, E., Nevo, R., Naveh, L., Levin-Zaidman, S., Kiss, V. et al. (2019). Chlorophyll catabolism precedes changes in chloroplast structure and proteome during leaf senescence. Plant Direct, 3(3), e00127. [Google Scholar]
14. Sakuraba, Y., Rahman, M. L., Cho, S. H., Kim, Y. S., Koh, H. J. et al. (2013). The rice faded green leaf locus encodes protochlorophyllide oxidoreductase B and is essential for chlorophyll synthesis under high light conditions. The Plant Journal, 74(1), 122–133. [Google Scholar]
15. Kusaba, M., Ito, H., Morita, R., Iida, S., Sato, Y. et al. (2007). Rice NON-YELLOW COLORING1 is involved in light-harvesting complex II and grana degradation during leaf senescence. Plant Cell, 19(6), 1362–1375. [Google Scholar]
16. Sato, Y., Morita, R., Katsuma, S., Nishimura, M., Tanaka, A. et al. (2009). Two short-chain dehydrogenase/reductases, NON-YELLOW COLORING 1 and NYC1-LIKE, are required for chlorophyll b and light-harvesting complex II degradation during senescence in rice. The Plant Journal, 57(1), 120–131. [Google Scholar]
17. Morita, R., Sato, Y., Masuda, Y., Nishimura, M., Kusaba, M. (2009). Defect in non-yellow coloring 3, an α/β hydrolase-fold family protein, causes a stay-green phenotype during leaf senescence in rice. The Plant Journal, 59(6), 940–952. [Google Scholar]
18. Yamatani, H., Sato, Y., Masuda, Y., Kato, Y., Morita, R. et al. (2013). NYC4, the rice ortholog of Arabidopsis THF1, is involved in the degradation of chlorophyll-protein complexes during leaf senescence. The Plant Journal, 74(4), 652–662. [Google Scholar]
19. Tang, Y. Y., Li, M. R., Chen, Y. P., Wu, P. Z., Wu, G. J. et al. (2011). Knockdown of OsPAO and OsRCCR1 cause different plant death phenotypes in rice. Journal of Plant Physiology, 168(16), 1952–1959. [Google Scholar]
20. Wang, L., Xu, J., Nian, J. Q., Shen, N. W., Lai, K. K. et al. (2016). Characterization and fine mapping of the rice gene OsARVL4 regulating leaf morphology and leaf vein development. Plant Growth Regulation, 78, 345–356. [Google Scholar]
21. Ye, W. J., Hu, S. K., Wu, L. W., Ge, C. W., Cui, Y. T. et al. (2016). White stripe leaf 12 (WSL12), encoding a nucleoside diphosphate kinase 2 (OsNDPK2regulates chloroplast development and abiotic stress response in rice (Oryza sativa L.). Molecular Breeding, 36, 1–15. [Google Scholar]
22. Zhao, J., Fang, Y. X., Kang, S. J., Ruan, B. P., Xu, J. et al. (2014). Identification and characterization of a new allele for ZEBRA LEAF 2, a gene encoding carotenoid isomerase in rice. South African Journal of Botany, 95, 102–111. [Google Scholar]
23. Ge, C. W., Wang, L., Ye, W. J., Wu, L. W., Cui, Y. T. et al. (2017). Single-point mutation of an histidine-aspartic domain-containing gene involving in chloroplast ribosome biogenesis leads to white fine Stripe leaf in rice. Scientific Reports, 7(1), 1–12. [Google Scholar]
24. Rao, Y. C., Xu, N., Li, S. F., Hu, J., Jiao, R. et al. (2019). PE-1, Encoding heme oxygenase 1, impacts heading date and chloroplast development in rice (Oryza sativa L.). Journal of Agricultural and Food Chemistry, 67(26), 7249–7257. [Google Scholar]
25. Rao, Y. C., Yang, Y. L., Xu, J., Li, X. J., Leng, Y. J. et al. (2015). Early senescence 1 encodes a SCAR-like protein 2 that affects water loss in rice. Plant Physiology, 169(2), 1225–1239. [Google Scholar]
26. Singh, S., Giri, M. K., Singh, P. K., Siddiqui, A., Nandi, A. K. (2013). Down-regulation of OsSAG12-1 results in enhanced senescence and pathogen-induced cell death in transgenic rice plants. Journal of Biosciences, 38(3), 583–592. [Google Scholar]
27. Kothari, K. S., Dansana, P. K., Giri, J., Tyagi, A. K. (2016). Rice stress associated protein 1 (OsSAP1) interacts with aminotransferase (OsAMTR1) and pathogenesis-related 1a protein (OsSCP) and regulates abiotic stress responses. Frontiers in Plant Science, 7, 1057. [Google Scholar]
28. Mao, C. J., Lu, S. C., Lv, B., Zhang, B., Shen, J. B. et al. (2017). A rice NAC transcription factor promotes leaf senescence via ABA biosynthesis. Plant Physiology, 174(3), 1747–1763. [Google Scholar]
29. Leng, Y. J., Ye, G. Y., Zeng, D. L. (2017). Genetic dissection of leaf senescence in rice. International Journal of Molecular Sciences, 18(12), 2686. [Google Scholar]
30. Fang, L. K., Li, Y. F., Gong, X. P., Sang, X. C., Ling, Y. H. et al. (2010). Genetic analysis and gene mapping of a dominant presenescing leaf gene PSL3 in rice (Oryza sativa L.). Chinese Science Bulletin, 55, 2517–2521. [Google Scholar]
31. Fang, C., Zhang, H., Wan, J., Wu, Y., Li, K. et al. (2016). Control of leaf senescence by an MeOH-Jasmonates cascade that is epigenetically regulated by OsSRT1 in rice. Molecular Plants, 9(10), 1366–1378. [Google Scholar]
32. Huang, L., Sun, Q., Qin, F., Li, C., Zhao, Y. et al. (2007). Down-regulation of a SILENT INFORMATION REGULATOR2-related histone deacetylase gene, OsSRT1, induces DNA fragmentation and cell death in rice. Plant Physiology, 144(3), 1508–1519. [Google Scholar]
33. Asada, K. (1999). The water-water cycle in chloroplasts: Scavenging of active oxygens and dissipation of excess photons. Annual Review of Plant Physiology and Plant Molecular Biology, 50, 601–639. [Google Scholar]
34. Hong, C. Y., Hsu, Y. T., Tsai, Y. C., Kao, C. H. (2007). Expression of Ascorbate peroxidase 8 in roots of rice (Oryza sativa L.) seedlings in response to NaCl. Journal of Experimental Botany, 58(12), 3273–3283. [Google Scholar]
35. Garg, R., Jhanwar, S., Tyagi, A. K., Jain, M. (2010). Genome-wide survey and expression analysis suggest diverse roles of glutaredoxin gene family members during development and response to various stimuli in rice. DNA Research, 17, 353–367. [Google Scholar]
36. Rong, H., Tang, Y., Zhang, H., Wu, P., Chen, Y. et al. (2013). The stay-green rice like (SGRL) gene regulates chlorophyll degradation in rice. Journal of Plant Physiology, 170(15), 1367–1373. [Google Scholar]
37. Kong, Z. S., Li, M. N., Yang, W. Q., Xu, W. Y., Xue, Y. B. (2006). A novel nuclear-localized CCCH-type zinc finger protein, OsDOS, is involved in delaying leaf senescence in rice. Plant Physiology, 141(4), 1376–1388. [Google Scholar]
38. Liang, C. Z., Wang, Y. Q., Zhu, Y. N., Tang, J. Y., Hu, B. et al. (2014). OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. PNAS, 111(27), 10013–10018. [Google Scholar]
39. Zhang, H., Zhao, Y., Zhou, D. X. (2017). Rice NAD+-dependent histone deacetylase OsSRT1 represses glycolysis and regulates the moonlighting function of GAPDH as a transcriptional activator of glycolytic genes. Nucleic Acids Research, 45(21), 12241–12255. [Google Scholar]
40. Ansari, M. I., Chen, S. C. G. (2009). Biochemical characterization of gamma-aminobutyric acid (GABAPyruvate transaminase during rice leaf senescence. International Journal of Integrative Biology, 6(1), 27–32. [Google Scholar]
41. Ishizaki, T., Ohsumi, C., Totsuka, K., Igarashi, D. (2010). Analysis of glutamate homeostasis by overexpression of Fd-GOGAT gene in Arabidopsis thaliana. Amino Acids, 38(3), 943–950. [Google Scholar]
42. Foyer, C. H., Noctor, G. (2005). Redox homeostasis and antioxidant signaling: A metabolic interface between stress perception and physiological responses. Plant Cell, 17(7), 1866–1875. [Google Scholar]
43. Saed-Moucheshi, A., Shekoofa, A., Pessarakli, M. (2014). Reactive oxygen species (ROS) generation and detoxifying in plants. Journal of Plant Nutrition, 37(10), 1573–1585. [Google Scholar]
44. Lee, S., Kim, J. H., Yoo, E. S., Lee, C. H., Hirochika, H. et al. (2005). Differential regulation of chlorophyll a oxygenase genes in rice. Plant Molecular Biology, 57(6), 805–818. [Google Scholar]
45. Mittler, R. (2017). ROS are good. Trends in Plant Science, 22(1), 11–19. [Google Scholar]
46. Del Río, L. A. (2015). ROS and RNS in plant physiology: An overview. Journal of Experimental Botany, 66(10), 2827–2837. [Google Scholar]
47. Dhindsa, R. S., Plumbdhindsa, P., Thorpe, T. A. (1981). Leaf senescence: Correlated with increased levels of membrane permeability and lipid peroxidation, and decreased levels of superoxide dismutase and catalase. Journal of Experimental Botany, 32(3), 93–101. [Google Scholar]
48. Rejeb, K. B., Abdelly, C., Savouré, A. (2012). Proline, a multifunctional amino-acid involved in plant adaptation to environmental constraints. Biologie Aujourdhui, 206(4), 291–299. [Google Scholar]
49. Yang, X., Gong, P., Li, K. Y., Huang, F. D., Cheng, F. M. et al. (2016). A single cytosine deletion in the OsPLS1 gene encoding vacuolar-type H+-ATPase subunit A1 leads to premature leaf senescence and seed dormancy in rice. Journal of Experimental Botany, 67(9), 2761–2776. [Google Scholar]
Appendix
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools