 Open Access
Open Access
ARTICLE
Transcriptome and Metabolome Revealed the Mechanism of NtBRL3 Overexpression Tobacco (Nicotiana tabacum L. K326) in Response to Drought Stress
College of Tobacco Science of Guizhou University, Guizhou Key Laboratory for Tobacco Quality, Guiyang, 550025, China
* Corresponding Author: Yang Liu. Email:
(This article belongs to the Special Issue: Identification of Genetic/Epigenetic Components Responding to Biotic and Abiotic Stresses in Crops)
Phyton-International Journal of Experimental Botany 2023, 92(9), 2555-2576. https://doi.org/10.32604/phyton.2023.030301
Received 30 March 2023; Accepted 19 May 2023; Issue published 28 July 2023
Abstract
Drought has severely affected the yield and quality of commercial crops. The BRI1 family plays an important role in plant response to drought stress, and BRL3 gene plays an important role in the study of drought in Arabidopsis thaliana. In this study, NtBRL3 was constructed as a vector and genetically transformed to obtain ‘N. Tobacco K326’ overexpression of NtBRL3. The enzyme activities of transgenic tobacco and wild-type tobacco were measured and transcriptome and metabolome analyses were performed. The results showed that the antioxidant enzymes of transgenic tobacco were more active under drought conditions, and 85 significantly differentially metabolites and 106 significantly differentially expressed genes were identified in the metabolome and transcriptome analyses, respectively. Transgenic tobacco NtBRL3ox demonstrated an excessive accumulation of drought-related metabolites, sugars such as sucrose and maltotetraose, and amino acids such as proline, compared with WT. We discovered drought-related differential genes in the root transcriptome, among which LOX6, RD22, WSD1, CCD8, and UGT were key genes which play an important role in plant response to drought stress. Our results demonstrate that NtBRL3 overexpression in K326 enhances drought resistance in transgenic tobacco.Keywords
Supplementary Material
Supplementary Material FileDrought is the main factor restricting the growth and quality of agricultural crops, affecting crop yield in particular [1]. Climatic anomalies and hydrological extremes have triggered domino effects on plant growth, development, and distribution. For example, increased temperature and insufficient precipitation have elevated evapotranspiration; thus, plants are subjected to stress during long periods in water-limited environments (i.e., due to droughts) [2]. Drought adversely affects not only the yield and quality of tobacco, an economically important and geographically widespread crop, but also the income and well-being of tobacco farmers. Tobacco (Nicotiana tabacum L.) is an economic crop that plays a highly important role in the national economy, and it is also one of the main economic crops cultivated in Guizhou, China. Guizhou has the second largest tobacco-growing area in the country. In 2014, the planting area of K326 significantly increased, accounting for 23.72% of the total-planting area in China, and was second only to that of ‘Yunyan 87’ [3]. K326 is becoming a widely cultivated variety in Guizhou, China. K326 was bred from the pure line parent of MnNair30 × NC95, and was promoted as an excellent variety in 1989. The leaf structure of the first flue-cured tobacco is loose, orange, rich in oil, harmonious in chemical composition, rich in aroma, and pure in taste. But, under abiotic stress of drought, K326 showed lower yield, lower chemical composition and less obvious drought resistance [4], which would cause serious economic losses. Therefore, it is particularly important to study and improve the drought resistance of K326.
Brassinosteroids (BR) participate in the process of regulating abiotic stress resistance in plants, and in particular, they can significantly promote plant resistance to drought, cold, high temperature, and diseases; these properties are important to improve the yield and quality of crops [5,6]. Researchers have used various materials to enrich the ability of BR to develop drought resistance in plants. For example, Li et al. studied Xanthoceras sorbifolia seedlings and found that BR increased leaf water content, relative water content, soluble sugar content, soluble protein content, free proline content, ascorbic acid content, and glutathione content, as well as SOD, POD, CAT, and APX activities [7]. Anjum et al. reported that exogenous BR sprayed on maize at the tasseling stage after drought stress regulated enzymatic antioxidant activity and gas-exchange properties [8]. Chen et al. pointed out that abscisic acid (ABA) and BR applied to tall fescue under water stress decreased stomatal conductance, transpiration, and net photosynthetic rate (Pn), whereas significantly increased net photosynthetic rate (Pn) and water-use efficiency (WUE) [9]. Therefore, BR may improve the ecophysiological plant response to drought stress [10]. Studies have shown that EBR can enhance the activity of antioxidant enzymes and the content of antioxidant substances, reduce peroxidative damage, promote the development of root morphology and the accumulation of osmotic adjustment substance content, alleviate the growth inhibition of drought stress on tobacco seedlings, and enhance drought resistance [11].
Brassinosteroid insensitive 1 (BRI1) is a receptor that senses and the protein encoded by the BRI1 genes initiate brassinosteroids (BR) signaling [12,13]. Kinoshita et al. demonstrated, using a biotin-tagged photoaffinity castasterone (BPCS), that active BRs bind directly to BRI1 [14]. BRI1 has three homologues, namely BRI1-like1 (BRL1), BRI1-like2 (BRL2), and BRI1-like3 (BRL3). Caño-Delgado identified the following three family members of BRI1 in Arabidopsis: AtBRL1 (At1g55610), AtBRL2 (At2g01950), and AtBRL3 (At3g13380). They revealed that only BRL1 and BRL3 act as functional BR receptors and can rescue the bri1 mutation [15,16]. A sequence analysis performed via sequence alignment indicated that BRL1 is a close homologue of BRI1; BRL1 is functionally redundant with BRI1 in regulating BR signaling in Arabidopsis; and bri1-4 deficiency can be rescued by overexpressing BRL1 in the bri1-4 defective phenotype [17]. BRI1 family plays an important role in BR signal activation, BR signal transduction affects the expression of genes in downstream pathways, and affects and improves plant drought resistance through related genes [18,19]. Fàbregas et al. overexpressed BRL3 (BRL3ox), which led not only to longer roots than the wild type but also increased levels of the receptor in root vascular tissue. BRL3ox accumulates osmoprotective metabolites in roots and shoots under drought stress [20]. Both longer root systems and accumulation of osmoprotective metabolites contribute to plant drought tolerance. However, there are few studies on the molecular mechanism of its influence on plant response to drought stress.
Flue-cured tobacco is an important economic crop from which the leaves are harvested, and the influence of water availability on this crop is clear. In recent years, due to global warming and subsequent rapid changes, water resources in various parts of our country have depleted, and droughts have occurred. Drought has a serious impact on the growth and development of flue-cured tobacco, resulting in different degrees of reduction in yield and quality. There is no report on the drought resistance of the BRI1 family in K326, and existing studies mostly focus on Arabidopsis. In this study, K326 was genetically transformed for overexpression of NtBRL3 and subjected to drought stress treatment experiments. Using transcriptome technology, the molecular mechanism of metabolome K326 transgenic tobacco under drought stress was investigated, to receive scientific evidence and cultivate drought-resistant tobacco varieties.
The complete tobacco genome and annotation files were downloaded from the Sol Genome Network (SGN1) (https://solgenomics.net/organism/Nicotiana_tabacum/genome, accessed on 5 January 2021) [21]. The four BRI1 family gene sequences of Arabidopsis were downloaded from TAIR (https://www.arabidopsis.org/). The four BRI1 family gene sequences of Arabidopsis—AtBRL1 (accession number:AT1G55610.1), AtBRL2 (accession number:AT2G01950.1), AtBRL3 (accession number: AT3G13380.1), and AtBRI1 (accession number:AT4G39400.1)—were compared using TBtools to obtain the possible sequences in the target species [22]. Public data (https://web.expasy.org/compute_pi/) were used to predict the physicochemical characteristics of the amino acid sequences of the four identified BRI1 family sequences, including their molecular weight (MW) and theoretical isoelectric point. Subcellular localization was predicted using Cell-PLoc 2.0.
2.2 Phylogenetic Trees, Motif Distribution, and Gene Structure
A phylogenetic tree was constructed using the neighborhood connection method [23]. A maximum-likelihood phylogeny based on the MAFFT alignment was constructed using the PhyML software under the WAG evolution model. A phylogenetic tree was visualized using FIGTREE. Conserved domains in NtBRI1 were identified using MEME. The full-length amino acid sequences of NtBRI1 were aligned using MAFFT with default parameters [24]. MEME software was used to identify conserved motifs with the default parameters3. Exon/intron information for the NtBRI1 genes was extracted from the corresponding genome annotation database. The data were then plotted using MapInspect software4.
2.3 Construction of Plasmids and Transformation of Plants
Introduction of vector plasmids into Agrobacterium tumefaciens. The complete coding sequence of NtBRL3 was amplified using primers (forward primer: 50-CAGTCGTCTCACAACATGGCATCTTTTCCTCTTGT-30 and reverse primer: 50-CAGTCGTCTCATACATCAGCTGAGTGTTTGATACC-30) from the cDNA of N. Tobacco K326 and cloned into the HindIII and XbaI sites of the expression vector pSH737. Transgenic tobacco were generated by the Agrobacterium tumefaciens-mediated transformation of strain LBA4404.
Tobacco genetic transformation: Shake and culture the single strain of Agrobacterium tumefaciens containing the NtBRL3 expression vector selected in the above steps on a shaker, stop the culture when the OD600 of the bacterial solution reaches 0.5, centrifuge the bacterial solution, and resuspend it with the resuspension solution before use. Using leaves of sterile tobacco seedlings as explants, tobacco was genetically transformed by agrobacterium-mediated leaf disc transformation.
The K326 sample used in this study was provided by the College of Tobacco Science of Guizhou University/Guizhou Key Laboratory for Tobacco Quality, Guiyang, 550025, China. Seedlings of K326 (Wild-type, WT) and overexpressed BRL3 mutant strain (named NtBRL3ox, three transgenic tobacco plants used in drought experiment were selected from the same callus.) with consistent growth were selected for drought stress treatment. Tobacco seedlings were rinsed thrice with distilled water and transferred to potted plants. Once tobacco seedlings had grown stably to five leaves-leaf one-heart, they were treated with 20% PEG6000 for 7 d to simulate drought stress.
WT K326 expression pattern under drought stress: Tobacco seeds were sown on a special substrate for tobacco with the following conditions: 75% relative humidity, temperature 28°C during the day (16 h) and 20°C at night (8 h), and foating seedlings. About 80 days after transplanting, the leaves, stems and roots of 3 similar healthy plants were collected; With the emergence of the sixth leaf, the plants showing similar growth vigor were selected and subjected to stress conditions as drought (15% PEG6000). Tree replicates were used for drought stress treatment; samples were collected after 0, 2, 12, and 24 h of treatment. The samples were either immediately frozen in liquid nitrogen or stored in an ultra-low temperature freezer at −80°C for subsequent qRT-PCR analysis. The weight of each sample was controlled at around 0.5 g.
2.5 RNA Extraction and Illumina Sequencing
Roots of K326 (WT) and NtBRL3ox were selected as samples after drought treatment (0.5 g) for total RNA extraction using an RNA extraction kit; a Qubit 2.0 Fluorometer was used to measure RNA concentrations with high accuracy. A NEBNext® UltraTM RNA Library Prep Kit was used for library construction. After completing library construction using a Qubit 2.0 Fluorometer for preliminary quantification, the library, at 1.5 ng/µL, and an Agilent 2100 bioanalyzer were used to detect the insert size of the library. Fastp was used to filter the raw data, mainly to remove reads with adapters. When the N content in any sequencing reads exceeded 10% of the base number of the reads, the paired reads were removed. When the number of low-quality (Q = 20) bases contained in any sequencing reads exceeded 50% of the number of bases in the reads, the paired reads were removed. All subsequent analyses were performed using clean reads. The enrichment analysis was based on a hypergeometric test for KEGG. A hypergeometric distribution test was used to identify pathways based on gene ontology (GO) terms. The false discovery rate (FDR) was obtained using the Benjamini–Hochberg method to correct the probability of hypothesis testing (p value). The screening conditions for differential genes were |log2folds Change| ≥ 1 and FDR < 0.05.
2.6 Qualitative and Quantitative Analyses of Metabolites
The biological samples were freeze-dried using a vacuum freeze dryer (Scientz-100F). The roots of WT and NtBRL3ox plants after drought treatment were crushed using a mixer mill (MM 400, Retsch) with zirconia beads for 1.5 min at 30 Hz. A total of 50 mg lyophilized powder was dissolved with 1.2 mL of the 70% methanol solution, and vortexed 6 times for 30 s at 30 min intervals. After centrifugation at 12,000 rpm for 3 min, the extracts were filtered (SCAA-104, 0.22 μm pore size; ANPEL, Shanghai, China, http://www.anpel.com.cn/) before UPLC-MS/MS analysis.
Significantly regulated metabolites between the groups were identified using absolute Log2FC (|Log2FC| ≥ 1.0). The identified metabolites were annotated using the KEGG compound database (http://www.kegg.jp/kegg/compound/), and the annotated metabolites were mapped to the KEGG Pathway database (http://www.kegg.jp/kegg/pathway.html). The pathways to metabolites with significant modulations were mapped and used in the Metabolite Set Enrichment Analysis (MSEA); their significance was determined using the p value of the hypergeometric test.
2.7 Gene Validation and Expression Analysis
For the purpose of the gene validation and expression analysis, the DEGs were subjected to a quantitative real-time polymerase chain reaction (qRT-PCR). qRT-PCR was performed using a 7500 Fast Quantitative Real-folds PCR instrument (Applied Biosystems, Waltham, MA, USA) and SYBR Premix Ex Taq (TaKaRa) according to the manufacturer’s instructions. The synthesized cDNA was used for the analysis of transcript abundance using qRT-PCR and the primers (Supplemental Table S1). The relative levels of the transcripts were determined by normalizing the expression against actin transcript levels. Experiments were replicated three times.
3.1 Genome-Wide Identification and Phylogenetic Analysis of BRL3 Gene in N. tabacum
The following four BRI1 families were identified in tobacco K326: Nitab4.5_0000184g0090.1(NtBRI1), Nitab4.5_0000175g0250.1(NtBRL2-1), Nitab4.5_0001822g0020.1(NtBRL2-2), and Nitab4.5_0000294g0170.1(NtBRL3). For reference, see Supplemental Table S2. The results of the physicochemical analysis revealed differences in the amino acid sequence length, MW, and pI values of these NtBRI1 proteins. The length and MW of NtBRI1 were the greatest in this family (1214 nt and 132,679.66 DA, respectively). NtBRL3 was similar to NtBRI1, and NtBRL2-1 was similar to NtBRL2-2. The difference was more pronounced between NtBRI1 and NtBRL3 than between NtBRL2-1 and NtBRL2-2. The subcellular localization prediction analysis showed that the NtBRI1 family was present on the cell membrane (Table 1). A phylogenetic tree was constructed based on the twenty-two BRI1 proteins present in the five species (Arabidopsis, rice, tomato, eggplant, and maize). BRI1 proteins from different plant species were divided into five main parts (Fig. 1A). The phylogenetic analysis showed that the four genes of tobacco BRI1 were distributed in three sublines as follows: NtBRI1 was in Group II, similar to Arabidopsis, rice, and tomato; NtBRL2-1 and NtBRL2-2 were in Group III, similar to tomato, rice, and eggplant; NtBRL3 and AT3G13380.1 (AtBRL3), rice and tomato were located in Group I; potato, eggplant, and rice were in Group IV; and potato and eggplant constituted an independent branch in Group V in the evolutionary tree distribution. Previous research cloned AT3G13380.1 (AtBRL3), which was identified in Arabidopsis, and constructed a mutant, revealing the positive role of BRL3 in drought resistance [8]. Therefore, based on the result that NtBRL3 is similar to AT3G13380.1, genetic transformation was carried out for K326.
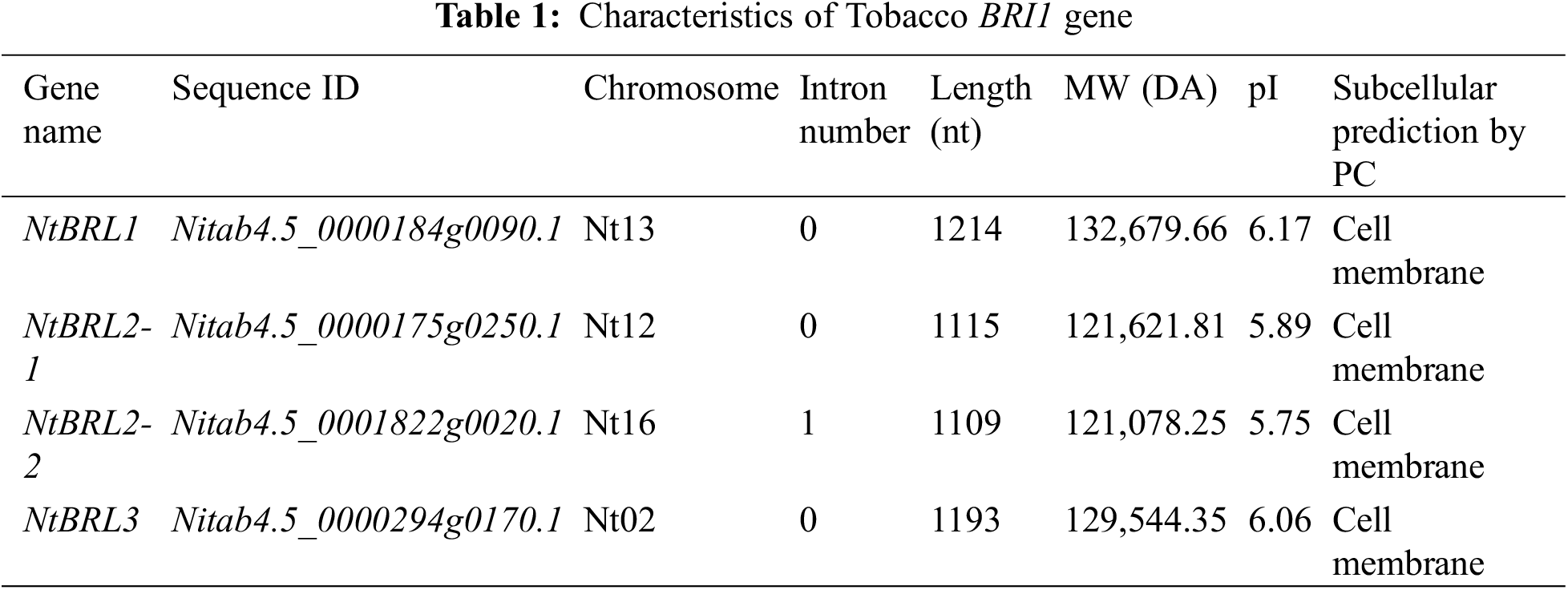
Figure 1: Bioinformatics analysis and expression patterns of tobacco gene families. (A) The rootless phylogenetic tree was constructed based on the gene sequences of BRI1 from Arabidopsis, tobacco, rice, tomato, potato and eggplant; each arc represents a group (a total of 5 groups). (B) Amino acid motifs in the NtBRI1 proteins (1–10) are represented by colored boxes. Black lines indicate relative protein lengths. (C) Exons and introns are indicated by yellow rectangles, and black lines, respectively. (D) Analysis of the 2000 bp upstream cis-acting element of NtBRI1 gene, different colors represent different types of cis-acting elements
3.2 Motif, Gene Structure and Expression Pattern Analysis of BRI1 Family Genes
A total of 10 conserved motifs (Motifs 1–10) were identified using the MEME suite. The gene structure showed that the distribution differences of the four NtBRI1 genes were mainly manifested at 0–400 bp; NtBRI1 has two additional Motif9 and one Motif8; NtBRL2-2 distributed two Motif7. The four genes had the same Motif at 400–1200 bp (Fig. 1B). The intron analysis revealed that only BRL2-2 contained one intron, while the remaining three NtBRI1 family genes showed a similar pattern of intronless organization (Fig. 1C). The structure of NtBRL3 without introns was the same as that of AT1G55610.1 (AtBRL1) and AT3G13380.1 (AtBRL3) without introns. Elements responsive to hormones, such as methyl jasmonate (MeJA), Gibberellin, and ABA, were identified in the promoter region 2000 bp upstream of the start codon of the BRI1 gene family (Fig. 1D). In particular, NtBRL3 found many cis-acting elements related to auxin response, such as light response, drought, low temperature, participation in defense, and stress response (Supplemental Sheet S1). NtBRL3 had no introns and contained many stress-related cis-acting elements, which forms the basis for further research.
The expression pattern of the NtBRI1 gene family in K326 was analyzed. The wild type K326 showed a higher expression of NtBRI1 in the root (Fig. 2). The expression levels of NtBRL2-1 and NtBRL2-2 decreased 2 h after drought stress treatment, especially in leaves. However, the expression of NtBRI1 and NtBRL3 increased in roots, and the expression reached the maximum at 12 h of treatment. Expression levels increased approximately twofold. The expression levels of NtBRI1 and NtBRL3 in stems and leaves were increased in leaves, but not as significantly as in roots.
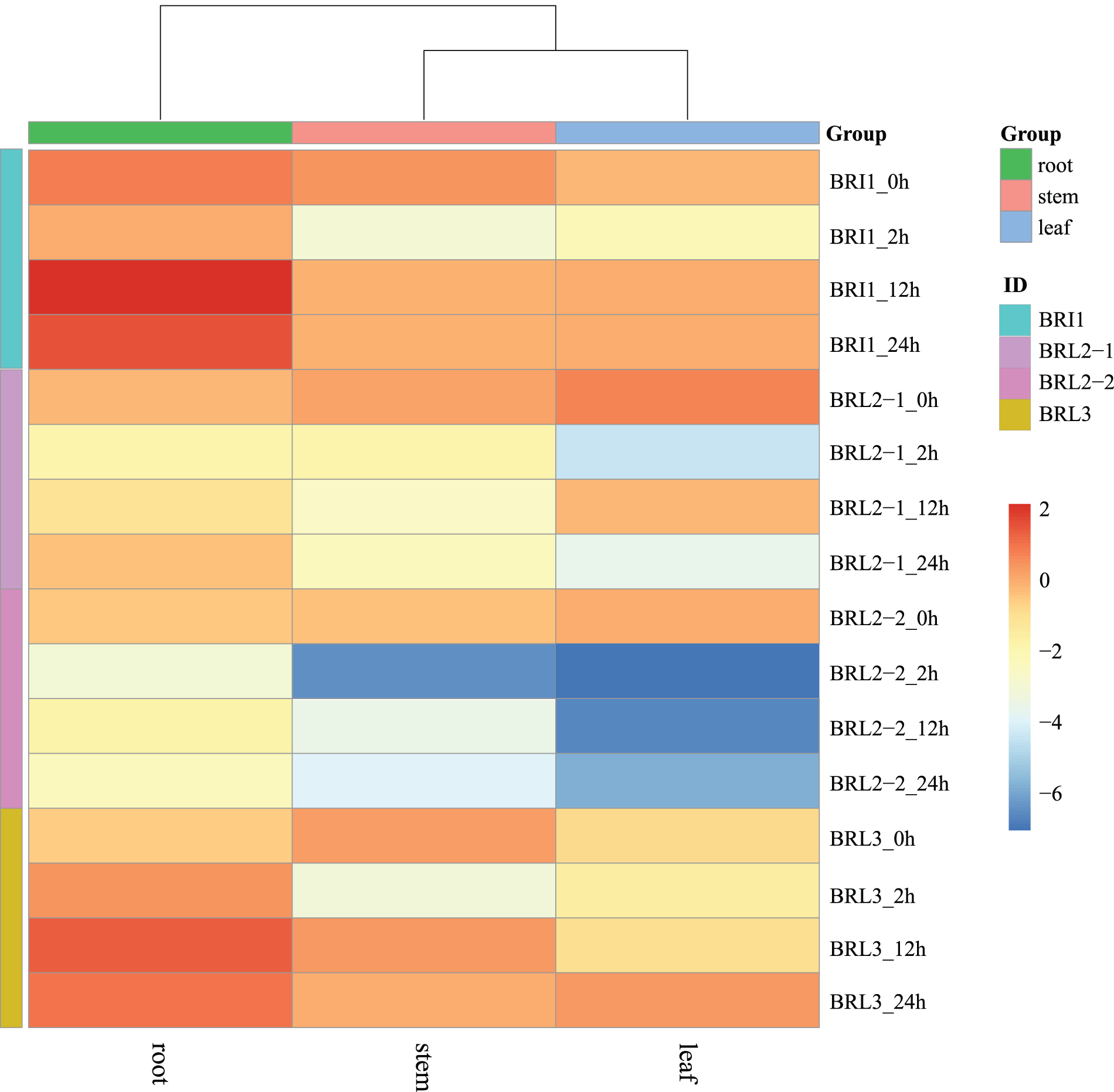
Figure 2: Expression pattern of BRI1 gene family in K326
3.3 Acquisition of Genetically Modified Materials and Drought Treatment
Genetically transformed plants of transgenic K326 were obtained (Fig. 3), the cells were cultured in the dark at 28°C for 2 d (Fig. 3A), the material was transferred to a culture medium containing antibiotics to grow callus (Fig. 3B), the shoots were cut off at about 2 cm in length to induce rooting (Fig. 3C), the plants selected for subsequent experiments were detected using PCR, the target band was obtained at 200 bp (Fig. 3D), photos of transgenic tobacco were captured before transplanting (Fig. 3E), and tobacco was transplanted and survived (Fig. 3F). From the phenotype point of view, there is no obvious change between the transgenic tobacco and the wild type. The drought stress tolerance of NtBRL3ox and WT plants with the same growth was tested after 7 d of drought in PEG6000. The growth and development of WT plants were severely inhibited, the leaves showed a certain degree of wilting, and the wilting leaves turned yellow, while the NtBRL3ox plants were less affected (Fig. 3G).

Figure 3: Genetic transformation of tobacco. (A) Dark combination of tobacco infected by Agrobacterium tumefaciens. (B) Transfer to medium containing Kan and Tim to grow callus. (C) Cut off shoots at about 2 cm in length to induce rooting. (D) PCR was performed on tobacco used in the drought experiment. (E) Photos of transgenic tobacco were captured before transplanting (upper transgenic, lower wild). (F) Tobacco transplanted and survived. (G) Phenotypic identification of NtBRL3ox and wild-type (WT) tobacco plants under drought treatment. (Left) NtBRL3ox and WT tobacco plants before drought stress. (Right) NtBRL3ox and WT plants at a similar status underwent PEG6000 treatment for 7 d
The enzyme activity of the samples was analyzed (Fig. 4). Under drought stress, the glutathione S-transferase (GST) activity of both groups increased. However, the GST activity of NtBRL3ox significantly increased, and the GST activity of NtBRL3ox was 1.51-fold higher than that of WT. The peroxidase (POD) activity of NtBRL3ox was 1.26-fold higher than that of WT. The results demonstrate that catalase (CAT) increased significantly in both NtBRL3ox and WT under drought stress; however, the increase in NtBRL3ox was 1.55-fold that of WT. Normally, the superoxide dismutase (SOD) activity in NtBRL3ox is slightly higher than that in WT. Under drought stress, the SOD activity of NtBRL3ox was 1.09-fold that of WT. This indicates that overexpression of NtBRL3 can improve the ROS scavenging ability of transgenic plants by increasing the enzyme activities of GST, POD, SOD, and CAT, thereby reducing ROS accumulation. These results indicated that overexpression of NtBRL3 enhanced the drought resistance of transgenic plants.
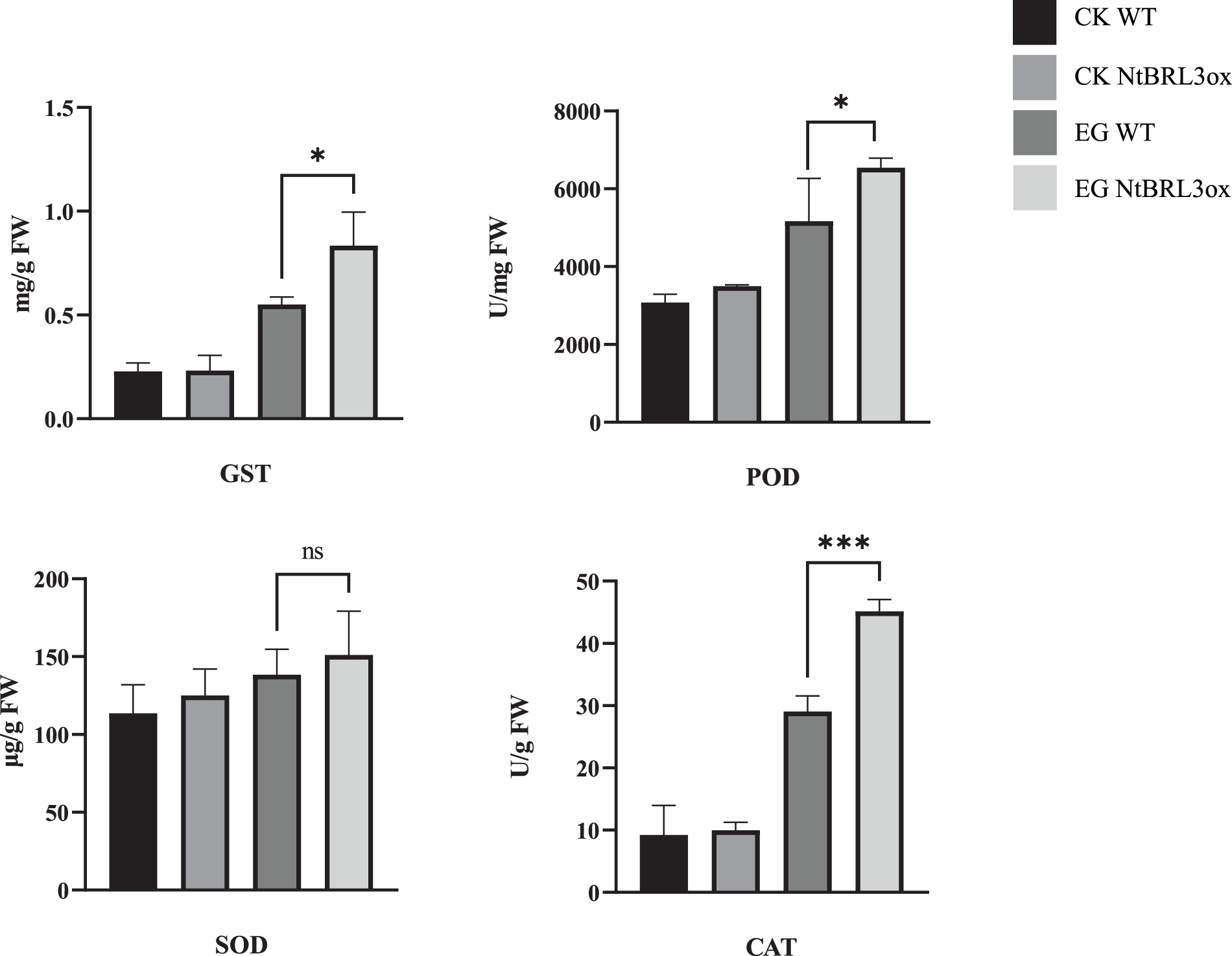
Figure 4: Enzyme activity of NtBRL3ox and WT drought stress experiments. Statistical analyses were performed by Student’s t-test and one-way ANOVA using SPSS 27.0 (SPSS Inc., Chicago, IL, USA). p = 0.05 was considered as statistically significant (*p < 0.05; ***p < 0.005; ns: none significantly). Graphs were prepared in Graphpad Prism (version 9.4.1). CK: Control Check; EG: Experimental Group
3.4 KEGG Functional Annotation and Enrichment Analysis of Differential Metabolites
To investigate whether NtBRL3 overexpression in K326 leads to drought resistance, we carried out metabolite profiling for BRL3ox plants and compared the results with those for WT in the same times-course drought experiments. The roots of NtBRL3ox and WT plants were analyzed for primary metabolites. A total of 528 differentially accumulated metabolites were detected, 85 of which were significantly different, and of which 33 were up-regulated and 52 were downregulated (Supplemental Sheet S2). Roots of NtBRL3ox plants exhibit a metabolic signature rich in free amino acids and sugars, which have been previously reported to be drought resistant [25,26]. Melezitose, maltose, maltotetraose, and sucrose were over-accumulated in NtBRL3ox buds compared with WT. The excessive accumulation of free amino acids such as proline, methionine, arginine, tyrosine, isoleucine, methionine, and asparagine was observed. This suggests that an increased amino acid content can serve as a precursor for stress-related metabolites, as well as an osmoregulatory source and energy fuel in plants [27]. This suggests that the NtBRL3 receptor facilitates the initiation of these metabolites [28].
The annotation results of KEGG with significant differential metabolites (DAMs) in the roots were classified according to pathway type. Metabolites with significant differences were mainly found in specific metabolic categories, among which the top four were metabolic pathways (44, 91.67%), the biosynthesis of secondary metabolites (20, 41.67%), the biosynthesis of amino acids (11, 22.92%), and the biosynthesis of cofactors (11, 22.92%). Apart from this, other highly differentially significant metabolites were concentrated in the vitamin B6 metabolism (3, 6.25%); tyrosine metabolism (4, 8.33%); pyrimidine metabolism (6, 12.5%); glyoxylate and dicarboxylate metabolism (5, 10.42%); glycine, serine, and threonine metabolism (6, 12.5%); glucosinolate biosynthesis (6, 12.5%); arginine and proline metabolism (5, 10.42%); alanine, aspartate, and glutamate metabolism (5, 10.42%); 2-oxocarboxylic acid metabolism (9, 18.75%); and aminoacyl-tRNA biosynthesis (9, 18.75%) (Fig. 5A, Supplemental Sheet S3). According to the results of the differential metabolites, the top 20 paths were selected according to the p value ranking from small to large.
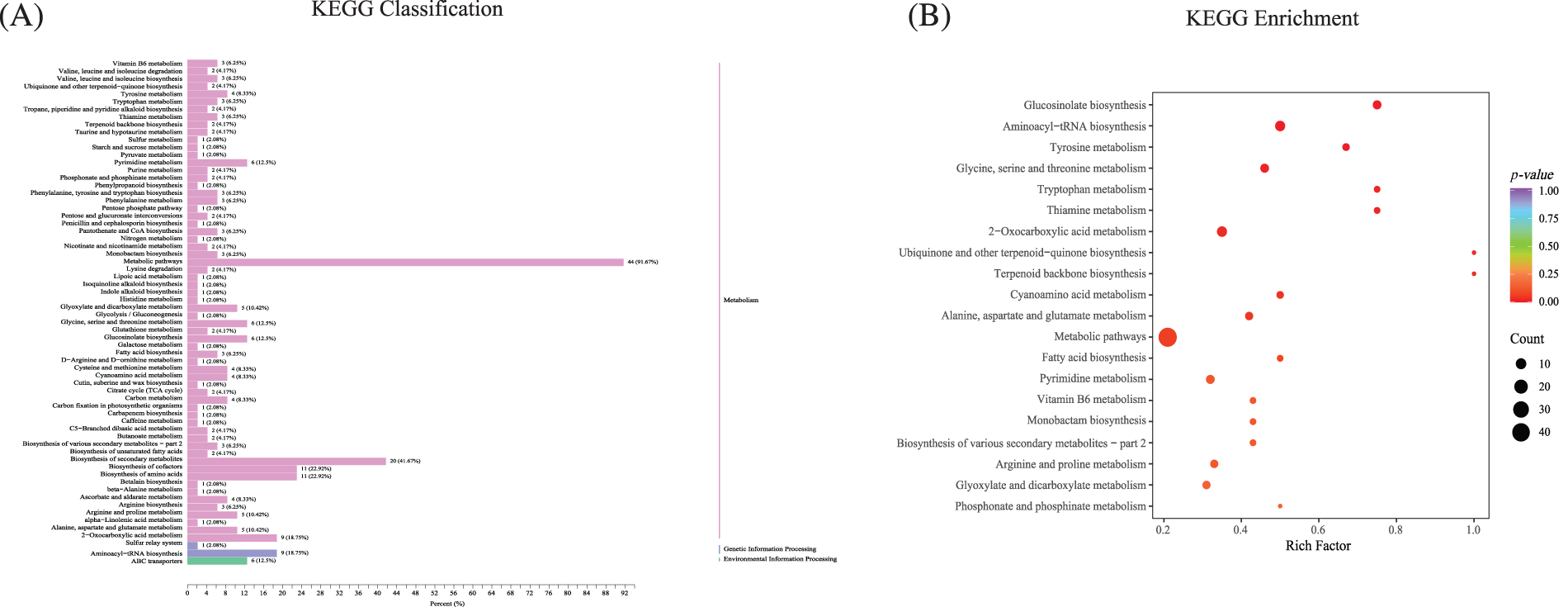
Figure 5: The annotation and enrichment results of KEGG with significant differential metabolites. (A) The annotation of KEGG of significant differential metabolites. (B) The KEGG enrichment of differential metabolites
A comprehensive analysis of differentially expressed genes and pathways with significant enrichment of differential metabolites found that NtBRL3ox can adapt to phenanthrene stress environments by increasing amino acid synthesis, increasing carbohydrate production and through other secondary metabolites. The KEGG enrichment analysis of differential metabolites found that the enriched pathways were mostly related to free amino acids, mainly in “glucosinolate biosynthesis”, “aminoacyl-tRNA biosynthesis”, “glycine, serine and threonine metabolism”, “Tryptophan metabolism”, “2-oxycarboxylic acid metabolism”, and “Arginine and proline metabolism”, which are mostly related to free amino acids, and the “Metabolism pathway” was the most enriched. (Fig. 5B, Supplemental Sheet S4).
3.5 Major Pathways Involved in NtBRL3ox
Differentially expressed genes (DEGs) in organisms perform biological functions through interactions, and pathway annotation analyses of differentially expressed genes are helpful for the further interpretation of gene functions. The genes were annotated in the KEGG database, and the number of differential genes contained in each KEGG pathway was counted (Fig. 6A). Environmental information processing category: “Plant hormone Signal Transduction” (3, 0.35%) and “MAPK Signaling Pathway-plant” (1, 0.14%). Genetic information processing category: “Protein Processing in the endoplasmic Reticulum” (2, 0.45%). In terms of the metabolism, the top three factors were “Metabolic pathway” (18, 0.39%), “Biosynthesis of secondary metabolites” (15, 0.65%), and “Glutathione metabolism” (3, 2.42%).
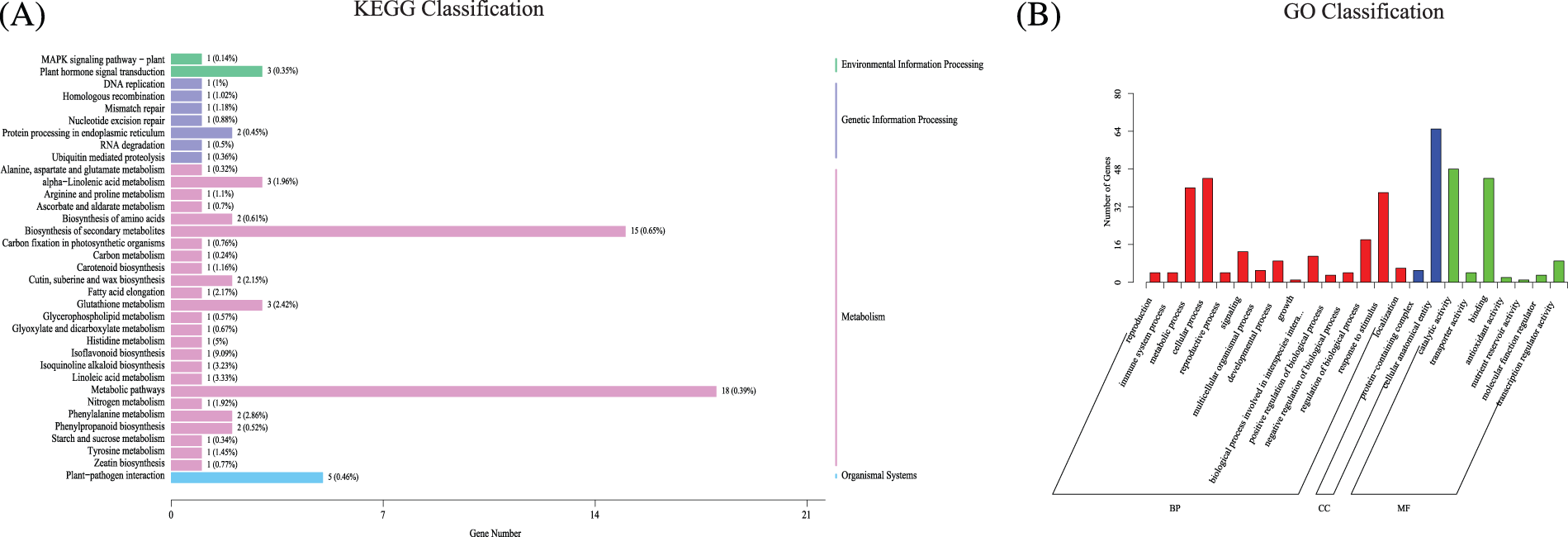
Figure 6: The annotation of significantly differential genes. (A) The annotation of KEGG of significant differential genes. (B) The annotation of GO of significant differential genes
According to a GO class analysis (Fig. 6B, Supplemental Sheet S5), 111 genes were annotated in the molecular function category. “Catalytic activity” and “Binding” were the most annotated groups, with 48 and 44 genes, respectively. A total of 70 genes were annotated in the category of cellular components, including “Cell anatomical entity” and “Protein−containing complex” which include 65 and 5 genes, respectively. A total of 226 genes were annotated in the biological process category, and 40, 44 and 38 genes were obtained in the three groups of “Metabolic process”, “Cellular process”, and “Stimulus response”.
KEGG and GO enrichment analyses were performed (Supplemental Sheets S6 and S7). The 20 KEGG pathways with the most significant enrichment levels were selected for analysis, and an enrichment scatter diagram was created. “Metabolism pathway” was the most enriched, and the number of genes enriched in “Biosynthesis of secondary metabolites” was significant. “Glutathione metabolism,” “alpha−Linolenic acid metabolism” and “Phenylalanine metabolism” were also enriched significantly. We also found that KEGG was enriched in “Phenylalanine metabolism,” “Tyrosine metabolism,” “Linoleic acid metabolism,” “Isoquinoline alkaloid biosynthesis,” “Isoflavone biosynthesis”, and “Cutin, suberine, and wax biosynthesis” (Fig. 7A, Supplemental Table S3). The 20 GO terms with the most significant enrichment were selected for analysis, and an enrichment scatter diagram was drawn (Fig. 7B, Supplemental Table S4). GO enrichment was mainly identified in “Phosphorelay relay signal transduction system”, “Cellular response to ethylene stimulus”, “ethylene−activated signaling pathway”, “Carbon–carbon lyase activity”, and “Carboxylate ester hydrolase activity”.
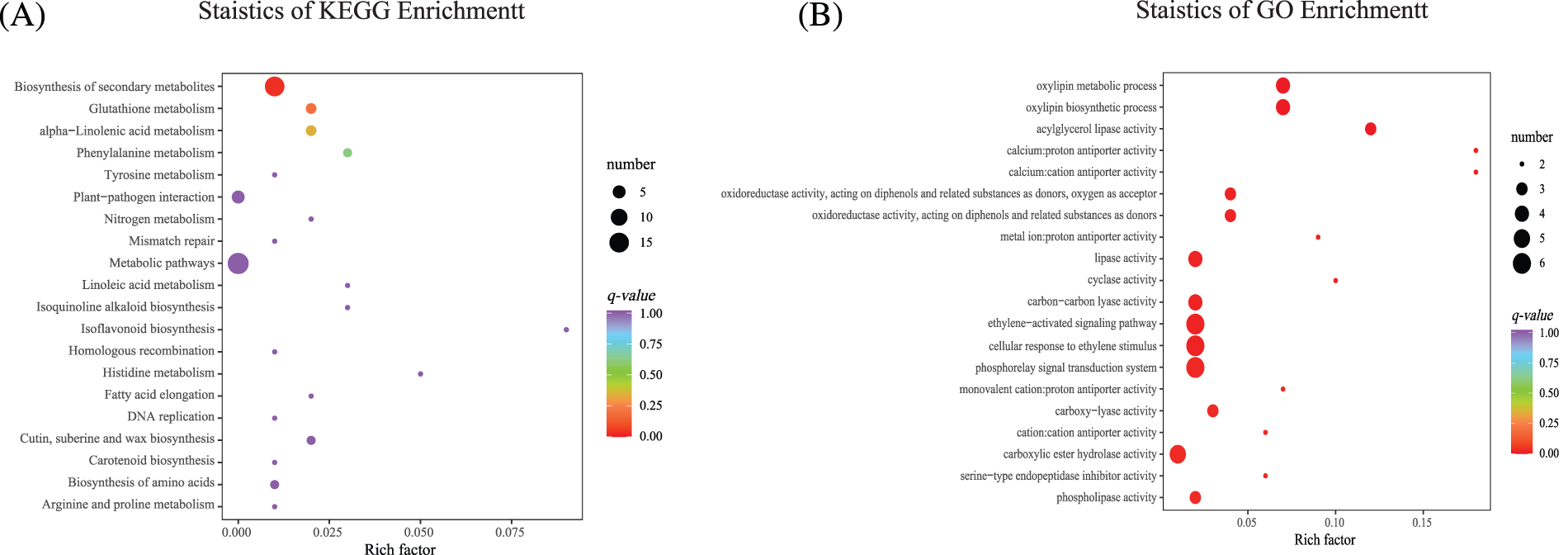
Figure 7: KEGG and GO enrichment analyses. (A) The 20 KEGG pathways with the most significant enrichment. (B) The 20 GO terms with the most significant enrichment
3.6 Expression Analysis of Drought Stress Key Genes
A total of 106 significantly different genes were found in NtBRL3ox compared to the WT analysis. In total, 49 DEGs in NtBRL3ox were upregulated, and 57 DEGs were downregulated in response to drought (Supplemental Sheet S8). Genes related to drought tolerance in DEGs were obtained, and these genes were involved in the response of plants to drought stress or induced expression by drought (Supplemental Table S5). To analyze the gene differences between NtBRL3ox and WT under drought stress, we identified 20 differentially expressed drought-related genes (p < 0.005). Some of these genes are involved in the response of plants to drought stress or are induced by drought, including Nitab4.5_0000848g0010 (glutathione S-transferase L3, GST) [29], Nitab4.5_0002227g0020 (phospholipase A1, PLA) [30], Nitab4.5_0001569g0130 (3-ketoacyl-CoA synthase 5, KCS) [31], Nitab4.5_0001937g0010 (linoleate 9S-lipoxygenase, LOX) [32], Nitab4.5_0000250g0250 (BURP domain protein RD22-like isoform X2, RD22) [33], Nitab4.5_0004977g0070 (cytochrome P450 86B1-like, alkane hydroxylase, MAH1) [34], Nitab4.5_0000185g0210 (shikimate O-hydroxycinnamoyltransferase-like, HCT) [35], Nitab4.5_0000638g0170 (O-acyltransferase, WSD1) [36], novel.12751 (wall-associated receptor kinase-like 20, WAK) [37] and novel.98 (carotenoid-cleaving dioxygenase 8, CCD8) [38].
The response of plants to drought stress is a process regulated by multiple genes; therefore, we analyzed and identified some key drought-related genes. We found that four genes (Nitab4.5_0001401g0130, Nitab4.5_0003727g0010, novel.3425, Nitab4.5_0001937g0010) were significantly up-regulated in response to JA. The 13-lipoxygenase LOX6 (Nitab4.5_0001937g0010), which is required for the synthesis of 12-OPDA, was expressed twofold in NtBRL3ox. Two RD22 (Nitab4.5_0000250g0250, Nitab4.5_0001403g0020) and WSD1 (Nitab4.5_0000638g0170) were found to be associated with ABA-induced responses [39,40], with significant upregulation by twofold. We also identified carotenoid cleavage dioxygenase (CCD8) (novel.98), which is related to the carotene cleavage dioxygenase process involved in strigolactone (SL) biosynthesis; it was found to be significantly upregulated twofold. Studies have demonstrated the role of UGT in plant stress defense mechanisms [41]. The bZIP transcription factor can combine with the UGT promoter to increase the transcription level of UGT. Two UGT (Nitab4.5_0000725g0120, novel.15930) were significantly upregulated by 2.7-fold and 1.8-fold in NtBRL3ox, and three bZIP transcription factors were positively expressed. In addition, we also detected glutathione S-transferase (GST). The expression of GST (Nitab4.5_0000848g0010) in NtBRL3ox was significantly upregulated, by about twofold. Overexpression of BRL3 enhanced the up-regulation of the above-mentioned genes related to various hormone-induced responses, and these genes were responsible for the enhanced drought resistance of transgenic tobacco.
According to a p value of <0.05 (Supplemental Sheet S9), transcription factors with significantly different expressions in different combination groups were identified (Supplemental Table S6). A transcriptome data analysis showed twenty-three AP2/ERF-ERF families, of which three were up-regulated and twenty were downregulated. The MYB, WRKY, and bHLH families responded significantly to drought stress, and most of them were downregulated. Among them, nine of the NAC family were upregulated. bHLH are negative regulators of proline biosynthesis, and we found that six bHLH were downregulated in NtBRL3ox.
3.7 Correlation Analysis of Transcriptome and Metabolome and Their Effects on Physiological Characteristics of Tobacco
To further determine the correlation between DEGs and DAMs, Pearson’s correlation analysis (r > 0.8 or −0.8; p value < 0.05) was performed (Fig. 8, Supplemental Sheet S10). UDP-glycosyltransferase (novel.15930, Nitab4.5_0000725g0120), abscisic acid and environmental stress-inducible protein TAS14-like (Nitab4.5_0000136g0200), BURP domain protein RD22-like (Nitab4.5_0004.5_, Nitab4.5_0000250g0250), and cytochrome P450 86B1-like (Nitab4.5_0004977g0070) were significantly associated with sucrose. UDP-glycosyltransferase (novel.15930, Nitab4.5_0000725g0120), BURP domain protein RD22-like (Nitab4.5_0001403g0020,Nitab4.5_0000250g0250), P450 86B1-like (Nitab4.5_0004977g0070), asparagine synthetase (novel.14492), and wall-associated receptor kinase-like (novel.12751) were significantly associated with maltose. UDP-glycosyltransferase (novel.15930,Nitab4.5_0000725g0120), abscisic acid, environmental stress-inducible protein TAS14-like (Nitab4.5_0000136g0200), and wall-associated receptor kinase-like (novel.12751) were significantly associated with melezitose. Except for maltotetraose, UDP-glycosyltransferase (Nitab4.5_0000725g0120) was significantly positively correlated with all saccharides.
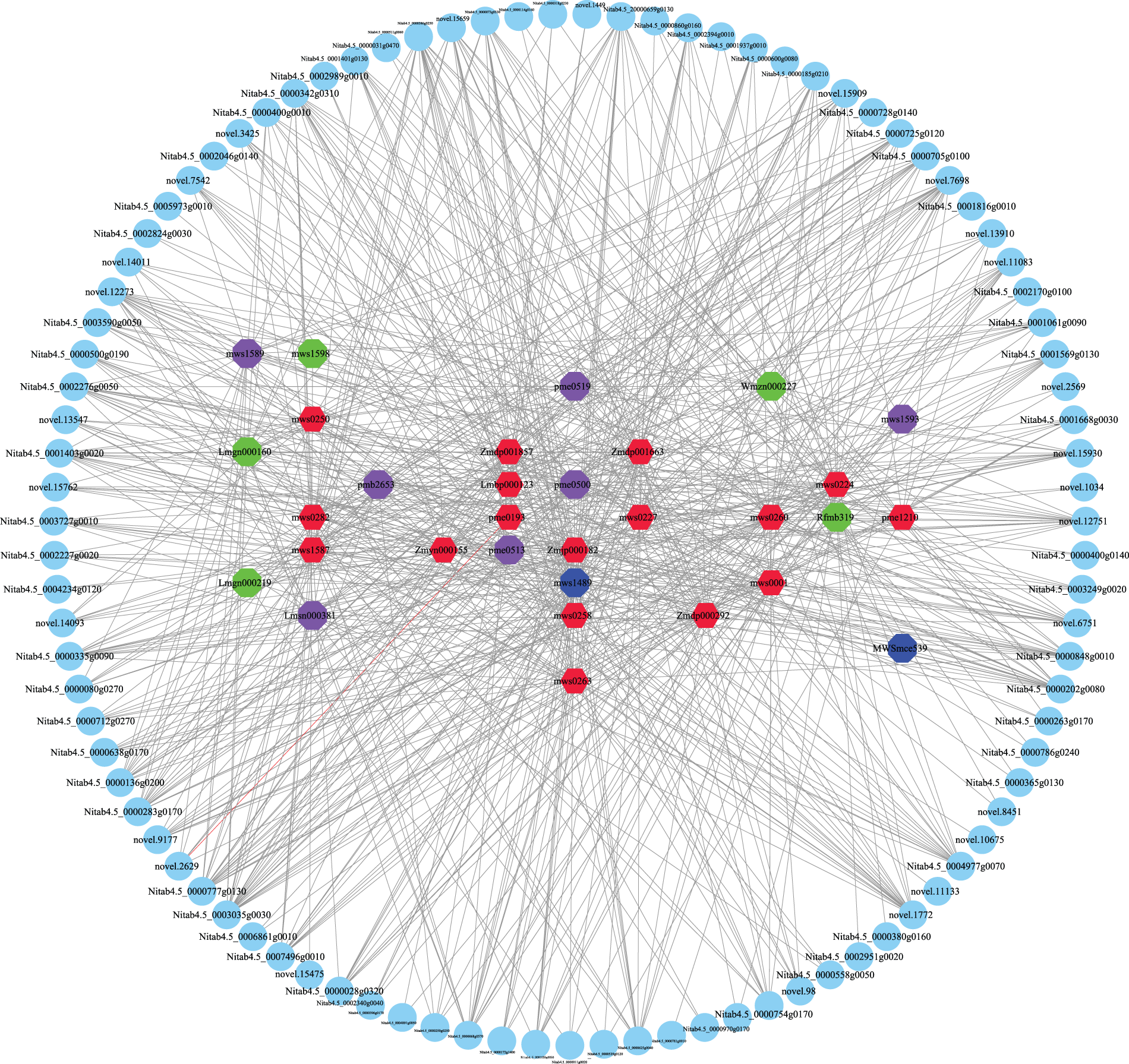
Figure 8: Schematic diagram of the correlation between transcriptome and metabolome
Cytochrome P450 86B1-like (Nitab4.5_0004977g0070), shikimate O-hydroxycinnamoyltransferase-like (Nitab4.5_0000185g0210), BURP domain protein RD22-like (Nitab4.5_0000250g0250, Nitab4.5_0001403g0020), carbonic anhydrase (novel.11083), carotenoid cleavage dioxygenase 8 (novel.98), 3-ketoacyl-CoA synthase (Nitab4.5_0001569g0130), asparagine synthase (novel.14492), and linoleate 9S-lipoxygenase (Nitab4.5_0001937g0010) were significantly positively correlated with proline. Flavin-dependent oxidoreductase (novel.13910), isoflavone 2’-hydroxylase-like (Nitab4.5_0003035g0030), and glutathione S-transferase (Nitab4.5_0000848g0010) were significantly positively correlated with glutamine. Glutathione S-transferase (Nitab4.5_0000848g0010), isoflavone 2′-hydroxylase-like (Nitab4.5_0003035g0030), cellulose synthase A catalytic subunit (Nitab4.5_0000342g0310), asparagine synthase (novel.14492), BURP domain protein RD22-like (Nitab4.5_0001403g0020, Nitab4.5_0000250g0250), cytochrome P450 86B1-like (Nitab4.5_0004977g0070), and wall-associated receptor kinase-like (novel.12751) were significantly positively correlated with methionine. Isoflavone 2′-hydroxylase-like (Nitab4.5_0003035g0030), glutathione S-transferase (Nitab4.5_0000848g0010), cellulose synthase A (Nitab4.5_0000342g0310), asparagine synthase (novel.14492), BURP domain protein RD22-like (Nitab4.5_0001403g0020, Nitab4.5_0000250g0250), cytochrome P450 86B1-like (Nitab4.5_0004977g0070), wall-associated receptor kinase-like (novel.12751), and UDP-glucosyltransferase (Nitab4.5_0000725g0120, novel.15930) were significantly positively correlated with arginine. Carotenoid cleavage dioxygenase 8 (novel.98), hydroperoxide dehydratase (novel.3425), shikimate O-hydroxycinnamoyltransferase-like (Nitab4.5_0000185g0210), cytochrome P450 86B1-like (Nitab4.5_0004977g0070), hydroperoxide dehydratase (Nitab4.5_0003727g0010), 3-ketoacyl-CoA synthase (Nitab4.5_0001569g0130), BURP domain protein RD22-like (Nitab4.5_0001403g0020 and Nitab4.5_0000250g0250), and linoleate 9S-lipoxygenase (Nitab4.5_0001937g0010) were significantly positively correlated with asparagine.
Among them, we noticed that the downregulation of negative regulators of metabolites, such as bHLH, likely relieves the restriction of proline accumulation. Other results may indicate that the accumulation of metabolites interacts with the expression of genes, and the up-regulated expression of these genes positively regulates the accumulation of metabolites, which is the reason for the increased drought resistance of NtBRL3ox.
3.8 Validation through qRT-PCR
The L25 gene was used as an internal reference gene to standardize the expression level of each gene. The original data were log2 transformed and compared to the qRT-PCR data (Supplemental Figure S1). The expression trends of qRT-PCR and RNA-seq sequencing were consistent; thus, the RNA-seq data were deemed to be reliable (Supplemental Figure S2). The expression of the NtBRL3 gene in NtBRL3ox was significantly higher than in WT, and the expression increased 2.5-fold (Supplemental Figure S3).
The development of human society and continuous population and consumption growth require a substantial increase in crop production to meet the growing demand for food. As an extreme weather event, drought is one of the main drivers of reduced crop yields worldwide and is expected to worsen in terms of its duration and intensity as a result of global climate change [42,43]. The exogenous application of BR compounds has been widely used in agriculture to promote growth under different drought stress conditions [44]. The protein encoded by the BRI1 family acts as the most active receptor for BR, and senses and initiates BR signaling [12]. In this study, the BRI1 family in K326 was systematically analyzed, a BRL3 overexpression vector was constructed, and K326 was genetically modified. The response of BRL3 to drought stress was analyzed through drought simulation experiments to discover drought candidate genes that can be used to screen drought-resistant K326 varieties.
4.1 BRI1 Family Characteristics
Four NtBRI1 genes (NtBRI1, NtBRL3, and two NtBRL2 genes) were identified in the K326 genome. BRL1 was not identified in K326. A phylogenetic tree analysis revealed that NtBRI1 is similar to At4G39400.1 (BRI1) (Fig. 1A). Although BRL1 was not identified in K326, NtBRI1 belongs to the same subline as Arabidopsis, rice, and tomato. AT1G55610.1 (AtBRL1) and AT3G13380.1 (AtBRL3) were very similar to NtBRL3, which indicated that AtBRL1, AtBRL3, and NtBRL3 may have evolved from the same ancestor. Previous studies have revealed that BRL1 and BRL3 have the highest homology, and the result of sequence analysis showed that they are 80% identical [14]. This finding suggests that NtBRL3 has a similar function to AtBRL3.
The motif of the BRI1 genes shows that there are differences between four genes at 0–400 (Fig. 1B). According to structural analysis (Fig. 1C), Nitab4.5_0001822g0020.1 (NtBRL2-2) in NtBRI1 contained one intron. None of the other members contained introns. An analysis of the cis-acting elements in the promoter sequence identified elements responsive to hormones (Fig. 1D), such as methyl jasmonate (MeJA), gibberellin, and ABA. Many functional elements related to auxin response (e.g., response to light, drought, and low temperature, and participation in defense and stress response) were found in the 2000 bp region upstream of the start codon in NtBRL3 (Supplemental Sheet S1). Intron-poor and intronless genes can be transcribed with little or no splicing, providing rapid responses to severe weather events [45]. Therefore, the structure of NtBRL3 with intronless and stress-related cis-acting elements plays an important role in its rapid response to drought.
The expression levels of NtBRL2-1 and NtBRL2-2 decreased 2 h after drought stress treatment, especially in leaves. However, the expression of NtBRI1 and NtBRL3 increased in roots, and maximum expression was reached at 12 h of treatment. The expression levels of NtBRI1 and NtBRL3 in stems and leaves showed a trend of first decreasing and then increasing; however, this trend was not as obvious as the increase in root expression after 12 and 24 h of treatment (Fig. 1E). This result indicates that drought stress could induce the expression of NtBRL3.
4.2 Accumulation of Osmoregulators in Roots of NtBRL3ox
Osmoregulation is considered an important process in plant adaptation to drought, as it helps to maintain tissue metabolic activity. Osmotic compounds synthesized under drought conditions include compatible solutes, such as amino acids (proline, aspartic acid, and glutamic acid), glycine betaine, sugars (fructan and sucrose), and cyclic alcohols (mannitol and pinitol) [46,47]. Sugars provide carbon and energy for the normal functioning of the cellular metabolism and regulate plant growth and development. Sugars and sugar alcohols have been widely accepted as osmoprotectants, which modulate osmoregulation, provide membrane protection, and scavenge toxic ROS against various stressors [48]. Analysis of the metabolome showed that, compared to WT, an increased accumulation of soluble sugars such as sucrose, maltose, melezitose, and maltotetraose occurred in the root tissue of NtBRL3ox, and the accumulation of these soluble sugars was related to drought resistance [49,50]. The function of xylitol is currently unclear; however, our results revealed that xylitol was significantly accumulated under drought stress.
As an important osmotic regulator, free amino acids can act as osmotic regulators, helping to reduce osmotic potential, thus maintaining cell expansion. We found that both glutamine and proline were accumulated in excess. Glutamine is the main amino acid involved in proline synthesis, and the accumulation of proline in plants results in increased tolerance to drought [51,52]. Therefore, we hypothesize that an increased glutamine content favors an increased proline content which ameliorated the detrimental effects of NtBRL3ox under drought conditions. Compared to the control, cauliflower sprayed with proline and methionine showed less pronounced reductions in growth parameters, with a higher leaf area and chlorophyll content [53]. Arginine treatment of wheat grains improved plant growth and mitigated the deleterious effects of drought stress [54]. The results of this study revealed an excessive accumulation of proline, methionine, and arginine, which may provide a basis for this. In addition, Kusaka et al. found that asparagine accumulated at high levels in leaves and stems after drought stress in pearl millet (IP8210) [55]. A significant increase in NtBRL3ox asparagine content was also observed in our results, with the difference that accumulation was found in the roots.
4.3 Expression of Genes in NtBRL3ox
The enhanced drought resistance of NtBRL3ox may be due to the enhanced level of the BRL3 protein, which in turn increases the response of brassinosteroids to enhance BR signaling. The number of brassinosteroid-binding sites and the degree of response to brassinosteroids depend on the level of BRI1 protein [14]. The expression of NtBRL3 in NtBRL3ox is 2.5-fold that of WT (Supplemental Figure S3), which means that the signal of the BR signaling pathway is enhanced. In plants, JA has been shown to act synergistically with BR for defense [56]. JA and JA precursor 12-OPDA can promote the closure of Arabidopsis stomata, and the increase in the OPDA content is related to the decrease in the stomatal opening and the improvement of drought resistance [57]. 13-Lipoxygenase LOX6 (Nitab4.5_0001937g0010) is required for the synthesis of 12-OPDA and plays an important role in plant drought tolerance. When drought stress is applied, LOX6 is synthesized into 12-OPDA, promoting stomatal closure and enhancing drought tolerance. Through the interaction between signaling pathways, BR can cooperate with ABA signaling to regulate plant drought resistance [58]. While the transcription of RD22 mRNA is induced by endogenous ABA, its production is triggered by drought stress. Plant-specific RD22-like proteins may act as drought-responsive genes to ABA signaling [59,60]. We found that two of the RD22 (Nitab4.5_0000250g0250, Nitab4.5_0001403g0020) were significantly upregulated in NtBRL3ox; we also found that 5 MYB were upregulated. The transcription factor MYB binds to the cis-element in the RD22 promoter to cooperatively activate RD22 [61]. Wax synthase/acyl-CoA: diacylglycerol acyltransferase (WSD1), which plays a critical role in wax ester synthesis transgenic Camelina lines overexpressing WSD1, showed enhanced tolerance to drought stress [62]. In NtBRL3ox, the expression of WSD1 was increased twofold. Therefore, we speculate that the upregulated expression of the two genes of RD22 and WSD1 may be related to the enhancement of BR signalling and the influence of the ABA signal. A carotenoid-cleaving dioxygenase (CCD8) gene associated with strigolactone (SL) synthesis was identified to be significantly upregulated, and the study also found that CCD8 (novel.98) was upregulated under drought conditions and positively regulated drought tolerance [63,64]. The enhanced drought resistance of NtBRL3ox may be due to the enhanced BRL3 protein level, which increases the response of brassinosteroids and thus enhances BR signaling. BR signaling works synergistically with other hormones for defense against drought by influencing signaling with other hormones.
In addition, we identified two glycosyltransferase genes (UGTs) that were significantly upregulated and noted that three bZIP were upregulated. Studies on rice have shown that the glycosyltransferase gene UGT2 plays a role in the bZIP transcription factor in adversity stress [65].
4.4 Correlation between Transcriptome and Metabolome Data
The correlation analysis was performed to detect the linear relationships between DEGs and DAMs (Fig. 7, Supplemental Sheet S10). An integrated analysis of the transcriptome and metabolome was performed to explore the linkage between metabolites and genes under NtBRL3ox drought stress. The P450 86B1-like gene was positively correlated with multiple sugar and amino acid metabolites. UDP-glycosyltransferase was significantly positively correlated with all sugars, except maltotetraose. UDP-glycosyltransferase (UGT) includes various plant hormones and metabolites that respond to biotic and abiotic stresses [66]. Therefore, the high expression of the UDP-glycosyltransferase gene in NtBRL3ox under drought stress may be related to the upregulation of carbohydrate metabolites. The BURP domain protein RD22-like (RD22) genes were highly correlated with multiple sugar and amino acid metabolites, suggesting that RD22 genes may be involved in the regulation of abiotic stress responses. The expression of RD22 is known to enhance plant tolerance to abiotic stress [30]. Studies have shown that bHLH is a negative regulator of proline biosynthesis, and we found that the expression coefficients of six bHLH were negatively correlated. From the results, the proline content was found to be significantly up-regulated [67].
In summary, we transformed NtBRL3 in tobacco K326, analyzed the transcriptome and metabolome after drought stress, and based on this, analyzed the function of candidate genes as transgenes. Transgenic plants expressing NtBRL3 were more resistant to drought. A comparative metabolome analysis revealed an excessive accumulation of drought-related metabolites, sugars such as sucrose, maltose, and maltotetraose, and amino acids such as proline, glutamic acid, methionine, and arginine in transgenic tobacco NtBRL3ox. A comparative transcriptome analysis revealed that the expression levels of multiple drought-responsive genes induced by hormones such as JA, ABA, and SL were increased in NtBRL3ox transgenic tobacco. We speculate that the enhanced drought resistance of transgenic tobacco is due to the enhanced expression of drought-related transcription factors such as NAC, MYB, bHLH, and bZIP, which are involved in the activation of gene expression by hormones such as JA and ABA. On the other hand, the enhanced BR signaling pathway may also affect the signal transduction of hormones such as JA, ABA, and SL, and genes sensitive to these hormones were significantly upregulated. Among them, RD22 and UGT were positively correlated with metabolites. BR signaling may initiate metabolite accumulation through interactions with other hormones, and genes associated with these hormones further promote metabolite accumulation.
Funding Statement: This study was financially supported by the Science and Technology Project of Guizhou Tobacco Company (2021XM04), the Creation of “Tobacco T-DNA Activation Insertion Mutant Library and Screening of Important Trait Mutants” Project of Guizhou University Talent Introduction (Guizhou University Hezi [2013] 50).
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: J.Y. and Y.L. planned and designed the research, analyzed the data, and wrote the manuscript. J.Y. and T.P. studied the gene expression by qRT-PCR. J.Y. identified the gene family and analyzed gene structure, chromosome distribution, and gene duplication, and performed syntenic analysis. Y.S. and X.L. analyzed the evolutionary relationship of NtBRI1 in several different species. K.W. and L.W. completed the genetic transformation experiment. J.Y., K.W. and L.W. performed the drought treatment experiment. Y.L. supervised the research. J.T. and D.W. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: All data generated or analysed during this study are included in this published article and its supplementary information files.
Ethics Approval: None.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Yang, X., Lu, M., Wang, Y., Wang, Y., Liu, Z. et al. (2021). Response mechanism of plants to drought stress. Horticulturae, 7(3), 50. https://doi.org/10.3390/horticulturae7030050 [Google Scholar] [CrossRef]
2. Iqbal, M. S., Singh, A. K., Ansari, M. I. (2020). Effect of drought stress on crop production. In: Rakshit, A., Singh, H. B., Singh, A. K., Singh, U. S., Fraceto, L. (Eds.New frontiers in stress management for durable agriculture, pp. 35–47. Singapore: Springer Singapore. [Google Scholar]
3. Sun, J. P., Wu, Z. H., Li, X. J., Sun, H., Ding, Y. F. et al. (2016). Analysis of regional variation and major varieties of flue-cured tobacco planted in China in the twenty-first century. Chinese Tobacco Science, 37(3), 86–92. [Google Scholar]
4. Ding, X. D., Xiong, J. B., Zhou, Z. Y., Ye, Q., Xiao, J. X. (2013). The response of Yunyan87 and K326 to drought stress. Acta Agriculturae Universitatis Jiangxiensis, 35(3), 507–511. [Google Scholar]
5. Basit, F., Liu, J., An, J., Chen, M., He, C. et al. (2021). Brassinosteroids as a multidimensional regulator of plant physiological and molecular responses under various environmental stresses. Environmental Science and Pollution Research, 28(33), 44768–44779. https://doi.org/10.1007/s11356-021-15087-8 [Google Scholar] [PubMed] [CrossRef]
6. Wang, X., Gao, Y., Wang, Q., Chen, M., Ye, X. et al. (2019). 24-Epibrassinolide-alleviated drought stress damage influences antioxidant enzymes and autophagy changes in peach (Prunus persicae L.) leaves. Plant Physiology and Biochemistry, 135(1), 30–40. https://doi.org/10.1016/j.plaphy.2018.11.026 [Google Scholar] [PubMed] [CrossRef]
7. Li, K. R., Feng, C. H. (2011). Effects of brassinolide on drought resistance of Xanthoceras sorbifolia seedlings under water stress. Acta Physiologiae Plantarum, 33(4), 1293–1300. https://doi.org/10.1007/s11738-010-0661-0 [Google Scholar] [CrossRef]
8. Anjum, S. A., Wang, L. C., Farooq, M., Hussain, M., Xue, L. L. et al. (2011). Brassinolide application improves the drought tolerance in maize through modulation of enzymatic antioxidants and leaf gas exchange. Journal of Agronomy and Crop Science, 197(3), 177–185. https://doi.org/10.1111/j.1439-037X.2010.00459.x [Google Scholar] [CrossRef]
9. Chen, Z., Wang, Z., Yang, Y., Li, M., Xu, B. (2018). Abscisic acid and brassinolide combined application synergistically enhances drought tolerance and photosynthesis of tall fescue under water stress. Scientia Horticulturae, 228, 1–9. https://doi.org/10.1016/j.scienta.2017.10.004 [Google Scholar] [CrossRef]
10. Singh Sidhu, G. P., Bali, A. S. (2022). Chapter 10—Plant responses to drought stress: Role of brassinosteroids. In: Ahammed, G. J., Sharma, A., Yu, J. (Eds.Brassinosteroids in plant developmental biology and stress tolerance, pp. 201–216. Academic Press. [Google Scholar]
11. Khan, R., Ma, X., Hussain, Q., Asim, M., Iqbal, A. et al. (2022). Application of 2,4-epibrassinolide improves drought tolerance in tobacco through physiological and biochemical mechanisms. Biology, 11(8), 1192. https://doi.org/10.3390/biology11081192 [Google Scholar] [PubMed] [CrossRef]
12. Jiang, J., Zhang, C., Wang, X. (2013). Ligand perception, activation, and early signaling of plant steroid receptor brassinosteroid insensitive 1. Journal of Integrative Plant Biology, 55(12), 1198–1211. https://doi.org/10.1111/jipb.12081 [Google Scholar] [PubMed] [CrossRef]
13. Kinoshita, T., Caño-Delgado, A., Seto, H., Hiranuma, S., Fujioka, S. et al. (2005). Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature, 433(7022), 167–171. https://doi.org/10.1038/nature03227 [Google Scholar] [PubMed] [CrossRef]
14. Wang, Z. Y., Seto, H., Fujioka, S., Yoshida, S., Chory, J. (2001). BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature, 410(6826), 380–383. https://doi.org/10.1038/35066597 [Google Scholar] [PubMed] [CrossRef]
15. Caño-Delgado, A., Yin, Y., Yu, C., Vafeados, D., Mora-García, S. et al. (2004). BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development, 131(21), 5341–5351. https://doi.org/10.1242/dev.01403 [Google Scholar] [PubMed] [CrossRef]
16. Yin, Y., Wu, D., Chory, J. (2002). Plant receptor kinases: Systemin receptor identified. Proceedings of the National Academy of Sciences of the United States of America, 99(14), 9090–9092. https://doi.org/10.1073/pnas.152330799 [Google Scholar] [PubMed] [CrossRef]
17. Zhou, A., Wang, H., Walker, J. C., Li, J. (2004). BRL1, a leucine-rich repeat receptor-like protein kinase, is functionally redundant with BRI1 in regulating Arabidopsis brassinosteroid signaling. The Plant Journal, 40(3), 399–409. https://doi.org/10.1111/j.1365-313X.2004.02214.x [Google Scholar] [PubMed] [CrossRef]
18. Belkhadir, Y., Jaillais, Y. (2015). The molecular circuitry of brassinosteroid signaling. New Phytologist, 206(2), 522–540. https://doi.org/10.1111/nph.13269 [Google Scholar] [PubMed] [CrossRef]
19. Shang, Y., Dai, C., Lee, M. M., Kwak, J. M., Nam, K. H. (2016). BRI1-associated receptor kinase 1 regulates guard cell ABA signaling mediated by open stomata 1 in Arabidopsis. Molecular Plant, 9(3), 447–460. https://doi.org/10.1016/j.molp.2015.12.014 [Google Scholar] [PubMed] [CrossRef]
20. Fàbregas, N., Lozano-Elena, F., Blasco-Escámez, D., Tohge, T., Martínez-Andújar, C. et al. (2018). Overexpression of the vascular brassinosteroid receptor BRL3 confers drought resistance without penalizing plant growth. Nature Communications, 9(1), 4680. https://doi.org/10.1038/s41467-018-06861-3 [Google Scholar] [PubMed] [CrossRef]
21. Edwards, K. D., Fernandez-Pozo, N., Drake-Stowe, K., Humphry, M., Evans, A. D. et al. (2017). A reference genome for Nicotiana tabacum enables map-based cloning of homeologous loci implicated in nitrogen utilization efficiency. BMC Genomics, 18(1), 448. https://doi.org/10.1186/s12864-017-3791-6 [Google Scholar] [PubMed] [CrossRef]
22. Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H. et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Molecular Plant, 13(8), 1194–1202. https://doi.org/10.1016/j.molp.2020.06.009 [Google Scholar] [PubMed] [CrossRef]
23. Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M. et al. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 28(10), 2731–2739. https://doi.org/10.1093/molbev/msr121 [Google Scholar] [PubMed] [CrossRef]
24. Katoh, K., Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molecular Biology and Evolution, 30(4), 772–780. https://doi.org/10.1093/molbev/mst010 [Google Scholar] [PubMed] [CrossRef]
25. Wu, X., Cai, K., Zhang, G., Zeng, F. (2017). Metabolite profiling of barley grains subjected to water stress: To explain the genotypic difference in drought-induced impacts on malting quality. Frontiers in Plant Science, 8, 1547. https://doi.org/10.3389/fpls.2017.01547 [Google Scholar] [PubMed] [CrossRef]
26. Durand, M., Porcheron, B., Hennion, N., Maurousset, L., Lemoine, R. et al. (2016). Water deficit enhances C export to the roots in Arabidopsis thaliana plants with contribution of sucrose transporters in both shoot and roots. Plant Physiology, 170(3), 1460–1479. https://doi.org/10.1104/pp.15.01926 [Google Scholar] [PubMed] [CrossRef]
27. Li, X., Ma, X., Cheng, Y., Liu, J., Han, L. et al. (2021). Transcriptomic and metabolomic insights into the adaptive response of Salix viminalis to phenanthrene. Chemosphere, 262(167), 127573. https://doi.org/10.1016/j.chemosphere.2020.127573 [Google Scholar] [PubMed] [CrossRef]
28. Conrath, U., Pieterse, C. M. J., Mauch-Mani, B. (2002). Priming in plant-pathogen interactions. Trends in Plant Science, 7(5), 210–216. https://doi.org/10.1016/S1360-1385(02)02244-6 [Google Scholar] [PubMed] [CrossRef]
29. Vaish, S., Gupta, D., Mehrotra, R., Mehrotra, S., Basantani, M. K. (2020). Glutathione S-transferase: A versatile protein family. 3 Biotech, 10(7), 321. https://doi.org/10.1007/s13205-020-02312-3 [Google Scholar] [PubMed] [CrossRef]
30. Zhai, L., Zhu, X., Yang, S., Gu, C., Liu, P. et al. (2021). Constitutive expression of a chrysanthemum phospholipase Dα gene in Chrysanthemum morifolium enhances drought tolerance. Ornamental Plant Research, 1(1), 1–10. https://doi.org/10.48130/OPR-2021-0008 [Google Scholar] [CrossRef]
31. Gao, X., Zhang, Y., Zhang, H., Huang, J. (2022). A β-ketoacyl-CoA synthase OsCUT1 confers increased drought tolerance in rice. Rice Science, 29(4), 353–362. https://doi.org/10.1016/j.rsci.2021.12.009 [Google Scholar] [CrossRef]
32. Gai, Z., Wang, Y., Ding, Y., Qian, W., Qiu, C. et al. (2020). Exogenous abscisic acid induces the lipid and flavonoid metabolism of tea plants under drought stress. Scientific Reports, 10(1), 12275. https://doi.org/10.1038/s41598-020-69080-1 [Google Scholar] [PubMed] [CrossRef]
33. Wang, H., Zhou, L., Fu, Y., Cheung, M. Y., Wong, F. L. et al. (2012). Expression of an apoplast-localized BURP-domain protein from soybean (GmRD22) enhances tolerance towards abiotic stress. Plant, Cell & Environment, 35(11), 1932–1947. https://doi.org/10.1111/j.1365-3040.2012.02526.x [Google Scholar] [PubMed] [CrossRef]
34. Greer, S., Wen, M., Bird, D., Wu, X., Samuels, L. et al. (2007). The cytochrome P450 enzyme CYP96A15 is the midchain alkane hydroxylase responsible for formation of secondary alcohols and ketones in stem cuticular wax of Arabidopsis. Plant Physiology, 145(3), 653–667. https://doi.org/10.1104/pp.107.107300 [Google Scholar] [PubMed] [CrossRef]
35. Shadle, G., Chen, F., Srinivasa Reddy, M. S., Jackson, L., Nakashima, J. et al. (2007). Down-regulation of hydroxycinnamoyl CoA: Shikimate hydroxycinnamoyl transferase in transgenic alfalfa affects lignification, development and forage quality. Phytochemistry, 68(11), 1521–1529. https://doi.org/10.1016/j.phytochem.2007.03.022 [Google Scholar] [PubMed] [CrossRef]
36. Huang, B. L., Li, X., Liu, P., Ma, L., Wu, W. et al. (2019). Transcriptomic analysis of Eruca vesicaria subs. sativa lines with contrasting tolerance to polyethylene glycol-simulated drought stress. BMC Plant Biology, 19(1), 419. https://doi.org/10.1186/s12870-019-1997-2 [Google Scholar] [PubMed] [CrossRef]
37. Degu, H. (2020). Analysis of differentially expressed genes induced by drought stress in tef (Eragrostistef) root. Nigerian Journal of Biotechnology, 36(2), 167–187. https://doi.org/10.4314/njb.v36i2.17 [Google Scholar] [CrossRef]
38. Gao, J., Zhang, T., Xu, B., Jia, L., Xiao, B. et al. (2018). CRISPR/Cas9-mediated mutagenesis of carotenoid cleavage dioxygenase 8 (CCD8) in tobacco affects shoot and root architecture. International Journal of Molecular Sciences, 19(4), 1062. https://doi.org/10.3390/ijms19041062 [Google Scholar] [PubMed] [CrossRef]
39. Matus, J. T., Aquea, F., Espinoza, C., Vega, A., Cavallini, E. et al. (2014). Inspection of the grapevine BURP superfamily highlights an expansion of RD22 genes with distinctive expression features in berry development and ABA-mediated stress responses. PLoS One, 9(10), e110372. https://doi.org/10.1371/journal.pone.0110372 [Google Scholar] [PubMed] [CrossRef]
40. Patwari, P., Salewski, V., Gutbrod, K., Kreszies, T., Dresen-Scholz, B. et al. (2019). Surface wax esters contribute to drought tolerance in Arabidopsis. The Plant Journal, 98(4), 727–744. https://doi.org/10.1111/tpj.14269 [Google Scholar] [PubMed] [CrossRef]
41. Li, Y. J., Wang, B., Dong, R. R., Hou, B. K. (2015). AtUGT76C2, an Arabidopsis cytokinin glycosyltransferase is involved in drought stress adaptation. Plant Science, 236, 157–167. https://doi.org/10.1016/j.plantsci.2015.04.002 [Google Scholar] [PubMed] [CrossRef]
42. Li, Y., Ye, W., Wang, M., Yan, X. (2009). Climate change and drought: A risk assessment of crop-yield impacts. Climate Research, 39, 31–46. https://doi.org/10.3354/cr00797 [Google Scholar] [CrossRef]
43. Lesk, C., Rowhani, P., Ramankutty, N. (2016). Influence of extreme weather disasters on global crop production. Nature, 529(7584), 84–87. https://doi.org/10.1038/nature16467 [Google Scholar] [PubMed] [CrossRef]
44. Tanveer, M., Shahzad, B., Sharma, A., Khan, E. A. (2019). 24-Epibrassinolide application in plants: An implication for improving drought stress tolerance in plants. Plant Physiology and Biochemistry, 135(1), 295–303. https://doi.org/10.1016/j.plaphy.2018.12.013 [Google Scholar] [PubMed] [CrossRef]
45. Liu, H., Lyu, H. M., Zhu, K., van de Peer, Y., Cheng, Z. M. (2021). The emergence and evolution of intron-poor and intronless genes in intron-rich plant gene families. The Plant Journal, 105(4), 1072–1082. https://doi.org/10.1111/tpj.15088 [Google Scholar] [PubMed] [CrossRef]
46. Kaur, G., Asthir, B. (2017). Molecular responses to drought stress in plants. Biologia Plantarum, 61(4), 201–209. https://doi.org/10.1007/s10535-016-0700-9 [Google Scholar] [CrossRef]
47. Sanders, G. J., Arndt, S. K. (2012). Osmotic adjustment under drought conditions. In: Aroca, R. (Ed.Plant responses to drought stress: From morphological to molecular features, pp. 199–229. Berlin, Heidelberg: Springer. [Google Scholar]
48. Singh, M., Kumar, J., Singh, S., Singh, V. P., Prasad, S. M. (2015). Roles of osmoprotectants in improving salinity and drought tolerance in plants: A review. Reviews in Environmental Science and Bio/Technology, 14(3), 407–426. https://doi.org/10.1007/s11157-015-9372-8 [Google Scholar] [CrossRef]
49. Ibrahim, H. A., Abdellatif, Y. M. R. (2016). Effect of maltose and trehalose on growth, yield and some biochemical components of wheat plant under water stress. Annals of Agricultural Sciences, 61(2), 267–274. https://doi.org/10.1016/j.aoas.2016.05.002 [Google Scholar] [CrossRef]
50. Liang, Y., Wei, G., Ning, K., Li, M., Zhang, G. et al. (2021). Increase in carbohydrate content and variation in microbiome are related to the drought tolerance of Codonopsis pilosula. Plant Physiology and Biochemistry, 165, 19–35. https://doi.org/10.1016/j.plaphy.2021.05.004 [Google Scholar] [PubMed] [CrossRef]
51. Brugière, N., Dubois, F., Limami, A. M., Lelandais, M., Roux, Y. et al. (1999). Glutamine synthetase in the phloem plays a major role in controlling proline production. The Plant Cell, 11(10), 1995–2011. https://doi.org/10.1105/tpc.11.10.1995 [Google Scholar] [PubMed] [CrossRef]
52. Živanović, B., Milić Komić, S., Tosti, T., Vidović, M., Prokić, L. et al. (2020). Leaf soluble sugars and free amino acids as important components of abscisic acid—mediated drought response in tomato. Plants, 9(9), 1147. https://doi.org/10.3390/plants9091147 [Google Scholar] [PubMed] [CrossRef]
53. EL-Bauome, H. A., Abdeldaym, E. A., Abd El-Hady, M. A. M., Darwish, D. B. E., Alsubeie, M. S. et al. (2022). Exogenous proline, methionine, and melatonin stimulate growth, quality, and drought tolerance in cauliflower plants. Agriculture, 12(9), 1301. https://doi.org/10.3390/agriculture12091301 [Google Scholar] [CrossRef]
54. Hussein, H. A. A., Alshammari, S. O., Kenawy, S. K. M., Elkady, F. M., Badawy, A. A. (2022). Grain-priming with L-arginine improves the growth performance of wheat (Triticum aestivum L.) plants under drought stress. Plants, 11(9), 1219. https://doi.org/10.3390/plants11091219 [Google Scholar] [PubMed] [CrossRef]
55. Kusaka, M., Ohta, M., Fujimura, T. (2005). Contribution of inorganic components to osmotic adjustment and leaf folding for drought tolerance in pearl millet. Physiologia Plantarum, 125(4), 474–489. https://doi.org/10.1111/j.1399-3054.2005.00578.x [Google Scholar] [CrossRef]
56. Yang, J., Duan, G., Li, C., Liu, L., Han, G. et al. (2019). The crosstalks between jasmonic acid and other plant hormone signaling highlight the involvement of jasmonic acid as a core component in plant response to biotic and abiotic stresses. Frontiers in Plant Science, 10, 1349. https://doi.org/10.3389/fpls.2019.01349 [Google Scholar] [PubMed] [CrossRef]
57. Savchenko, T., Kolla, V. A., Wang, C. Q., Nasafi, Z., Hicks, D. R. et al. (2014). Functional convergence of oxylipin and abscisic acid pathways controls stomatal closure in response to drought. Plant Physiology, 164(3), 1151–1160. https://doi.org/10.1104/pp.113.234310 [Google Scholar] [PubMed] [CrossRef]
58. Gupta, A., Rico-Medina, A., Caño-Delgado, A. I. (2020). The physiology of plant responses to drought. Science, 368(6488), 266–269. https://doi.org/10.1126/science.aaz7614 [Google Scholar] [PubMed] [CrossRef]
59. Yamaguchi-Shinozaki, K., Shinozaki, K. (1993). The plant hormone abscisic acid mediates the drought-induced expression but not the seed-specific expression of rd22, a gene responsive to dehydration stress in Arabidopsis thaliana. Molecular & General Genetics MGG, 238(1–2), 17–25. https://doi.org/10.1007/BF00279525 [Google Scholar] [PubMed] [CrossRef]
60. Xu, H., Li, Y., Yan, Y., Wang, K., Gao, Y. et al. (2010). Genome-scale identification of Soybean BURP domain-containing genes and their expression under stress treatments. BMC Plant Biology, 10(1), 197. https://doi.org/10.1186/1471-2229-10-197 [Google Scholar] [PubMed] [CrossRef]
61. Abe, H., Urao, T., Ito, T., Seki, M., Shinozaki, K. et al. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. The Plant Cell, 15(1), 63–78. https://doi.org/10.1105/tpc.006130 [Google Scholar] [PubMed] [CrossRef]
62. Abdullah, H. M., Rodriguez, J., Salacup, J. M., Castañeda, I. S., Schnell, D. J. et al. (2021). Increased cuticle waxes by overexpression of WSD1 improves osmotic stress tolerance in Arabidopsis thaliana and Camelina sativa. International Journal of Molecular Sciences, 22(10), 5173. https://doi.org/10.3390/ijms22105173 [Google Scholar] [PubMed] [CrossRef]
63. Xia, X., Dong, H., Yin, Y., Song, X., Gu, X. et al. (2021). Brassinosteroid signaling integrates multiple pathways to release apical dominance in tomato. Proceedings of the National Academy of Sciences of the United States of America, 118(11), e2004384118. https://doi.org/10.1073/pnas.2004384118 [Google Scholar] [PubMed] [CrossRef]
64. Ha, C. V., Leyva-González, M. A., Osakabe, Y., Tran, U. T., Nishiyama, R. et al. (2014). Positive regulatory role of strigolactone in plant responses to drought and salt stress. Proceedings of the National Academy of Sciences of the United States of America, 111(2), 851–856. https://doi.org/10.1073/pnas.1322135111 [Google Scholar] [PubMed] [CrossRef]
65. Wang, T., Li, X. K., Liu, X., Yang, X. Q., Li, Y. J. et al. (2023). Rice glycosyltransferase gene UGT2 functions in salt stress tolerance under the regulation of bZIP23 transcription factor. Plant Cell Reports, 42(1), 17–28. https://doi.org/10.1007/s00299-022-02933-3 [Google Scholar] [PubMed] [CrossRef]
66. Rehman, H. M., Nawaz, M. A., Shah, Z. H., Ludwig-Müller, J., Chung, G. et al. (2018). Comparative genomic and transcriptomic analyses of Family-1 UDP glycosyltransferase in three Brassica species and Arabidopsis indicates stress-responsive regulation. Scientific Reports, 8(1), 1875. https://doi.org/10.1038/s41598-018-19535-3 [Google Scholar] [PubMed] [CrossRef]
67. Verma, D., Jalmi, S. K., Bhagat, P. K., Verma, N., Sinha, A. K. (2020). A bHLH transcription factor, MYC2, imparts salt intolerance by regulating proline biosynthesis in Arabidopsis. The FEBS Journal, 287(12), 2560–2576. https://doi.org/10.1111/febs.15157 [Google Scholar] [PubMed] [CrossRef]
Supplementary Materials
Supplemental Figure S1: The original data of the selected 10 genes were log2 transformed.
Supplemental Figure S2: Relative expression profile of genes by real-time PCR. Relative quantification was obtained through 2−(ΔΔCT) method using L25 as reference gene (p value < 0.05).
Supplemental Figure S3: Relative expression profile of NtBRL3 gene in NtBRL3ox and WT detected by real-time PCR (p value < 0.05).
Supplemental Sheet S1: Promoter region 2000 bp upstream of the cis-elements.
Supplementary Sheet S2: Differentially significant metabolites.
Supplementary Sheet S3: Differentially significant metabolite KEGG annotation.
Supplementary Sheet S4: Differential metabolite KEGG enrichment.
Supplemental Sheet S5: The number of GO annotations obtained.
Supplemental Sheet S6: Differentially significant metabolite KEGG annotation.
Supplemental Sheet S7: The 20 GO-Terms with the most significant enrichment.
Supplemental Sheet S8: Differentially significant genes.
Supplemental Sheet S9: Differential genes (p value < 0.05).
Supplemental Sheet S10: Correlation between DEGs and DAMs.
Supplemental Table S1: Genes and corresponding primers selected in qPCR.
Supplemental Table S2: NtBRI1 family sequences.
Supplemental Table S3: KEGG pathway annotation and number of DEGs in different comparison groups.
Supplemental Table S4: GO annotations and number of DEGs.
Supplemental Table S5: Drought-related candidate genes.
Supplemental Table S6: Transcription Factors family obtained.
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools