Integrative Machine Learning and Experimental Validation Identify MYBL2 as a Prognostic Biomarker and Therapeutic Target in Hepatocellular Carcinoma
Ya-Ling Yang1,#, Ying-Hsien Huang2,#, Hung-Yu Lin3,4,*
1 Department of Anesthesiology, Kaohsiung Chang Gung Memorial Hospital and Chang Gung University College of Medicine, Kaohsiung, Taiwan
2 Department of Pediatrics, Kaohsiung Chang Gung Memorial Hospital and Chang Gung University College of Medicine, Kaohsiung, Taiwan
3 Department of Post-Baccalaureate Medicine, College of Medicine, National Chung Hsing University, Taichung, Taiwan
4 Research Assistant Center, Show Chwan Memorial Hospital, Changhua, Taiwan
* Corresponding Author: Hung-Yu Lin. Email:  ,
, 
# These authors contributed equally to this work
(This article belongs to the Special Issue: Tumor Biomarkers for Diagnosis, Prognosis and Targeted Therapy)
Oncology Research https://doi.org/10.32604/or.2026.075284
Received 29 October 2025; Accepted 08 January 2026; Published online 26 January 2026
Abstract
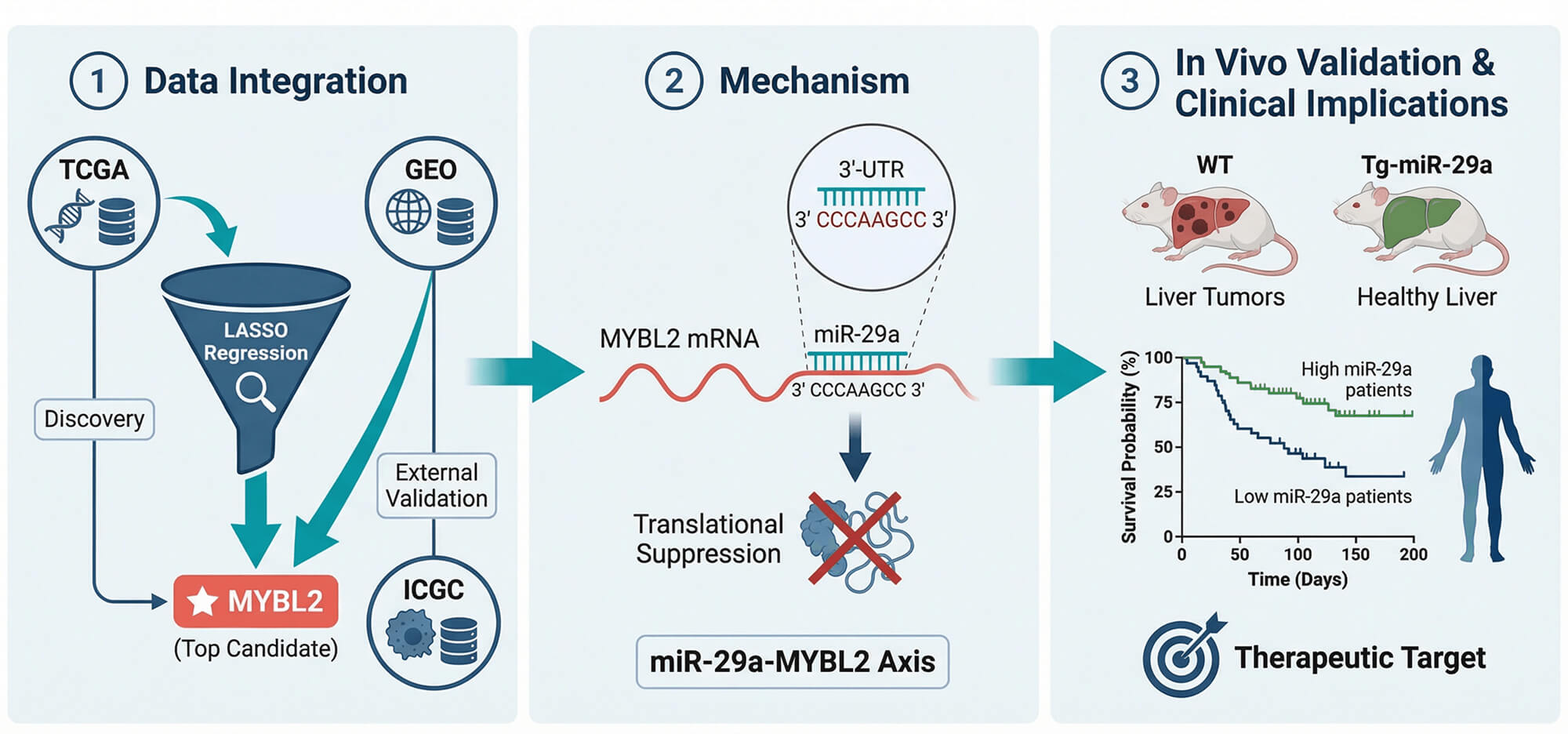
Background: Hepatocellular carcinoma (HCC) presents with poor treatment outcomes, creating an urgent need for novel biomarkers to improve diagnosis, prognosis, and precision medicine. While the MYB family of oncogenes is implicated in cancer, the role and regulatory mechanisms of its member, particularly MYB proto-oncogene like 2 (MYBL2), remain underexplored in HCC. Therefore, this study aimed to systematically validate the clinical significance of MYBL2, elucidate its functional role in tumor progression and drug sensitivity, and identify its upstream regulatory mechanisms using an integrative machine learning and experimental framework.
Methods: We applied an integrative pipeline combining LASSO-based feature selection on TCGA and GEO cohorts, single-cell transcriptomics, pharmacogenomic surveys, and CRISPR dependency screens. These computational approaches were complemented by
in vitro HepG2 assays, luciferase reporter tests, iTRAQ proteomics, and an
in vivo western diet/CCl
4 (WD/CCl
4) HCC model using miR-29a transgenic mice to investigate a putative regulatory axis.
Results: MYBL2 robustly discriminated tumor from normal liver (AUC = 0.968), and high expression was associated with adverse features, including higher grade, microvascular invasion, HBV positivity, nonresponse to TACE, and worse survival. A nomogram combining MYBL2 with AJCC stage improved 1-, 3-, and 5-year AUCs versus stage alone. MYBL2 correlated with proliferative biomarkers (AFP, MKI67, PCNA, BIRC5) and CRISPR knockout inhibited growth in most HCC lines. High MYBL2 expression was associated with greater sensitivity to sorafenib in pharmacogenomic screens and was linked to an immunosuppressive microenvironment and higher MSI. Mechanistically, miR-29a was shown to suppress MYBL2 translation by directly binding to its 3
′-UTR; this was validated
in vivo, where miR-29a transgenic mice were protected from WD/CCl
4-induced HCC, demonstrating reduced tumor burden, MYBL2 expression, and fibrosis. iTRAQ proteomics further confirmed MYBL2 as a top miR-29a–regulated protein.
Conclusions: MYBL2 is a potent diagnostic and prognostic biomarker in HCC that also predicts sorafenib sensitivity. Our findings establish a clear regulatory link where MYBL2 is a direct and functionally important target of the tumor-suppressive miR-29a. This positions MYBL2 as a tractable target for miR-29a-based therapeutic strategies, warranting clinical validation for patient stratification and treatment development in HCC.
Graphical Abstract
Keywords
Hepatocellular carcinoma; MYBL2; biomarker; prognosis; precision medicine; miR-29a; tumor microenvironment
 Open Access
Open Access,