Integrated Global Phosphoproteomic, Bioinformatic, and Machine Learning Analysis Reveals Regulatory Networks of TOP1, TOP2A, TOP2B, and C1orf35 in Hepatocellular Carcinoma (HCC)
Aktham Mestareehi1,2,*
1 Department of Applied Pharmaceutical Sciences and Clinical Pharmacy, Faculty of Pharmacy, Isra University, Amman, Jordan
2 Department of Pharmaceutical Sciences, Eugene Applebaum College of Pharmacy and Health Sciences, Wayne State University, Detroit, MI, USA
* Corresponding Author: Aktham Mestareehi. Email: 
(This article belongs to the Special Issue: Advances in Liver Cancer: Novel Therapeutics and Biomarkers for HCC and CCA)
Oncology Research https://doi.org/10.32604/or.2026.073745
Received 24 September 2025; Accepted 12 January 2026; Published online 03 February 2026
Abstract
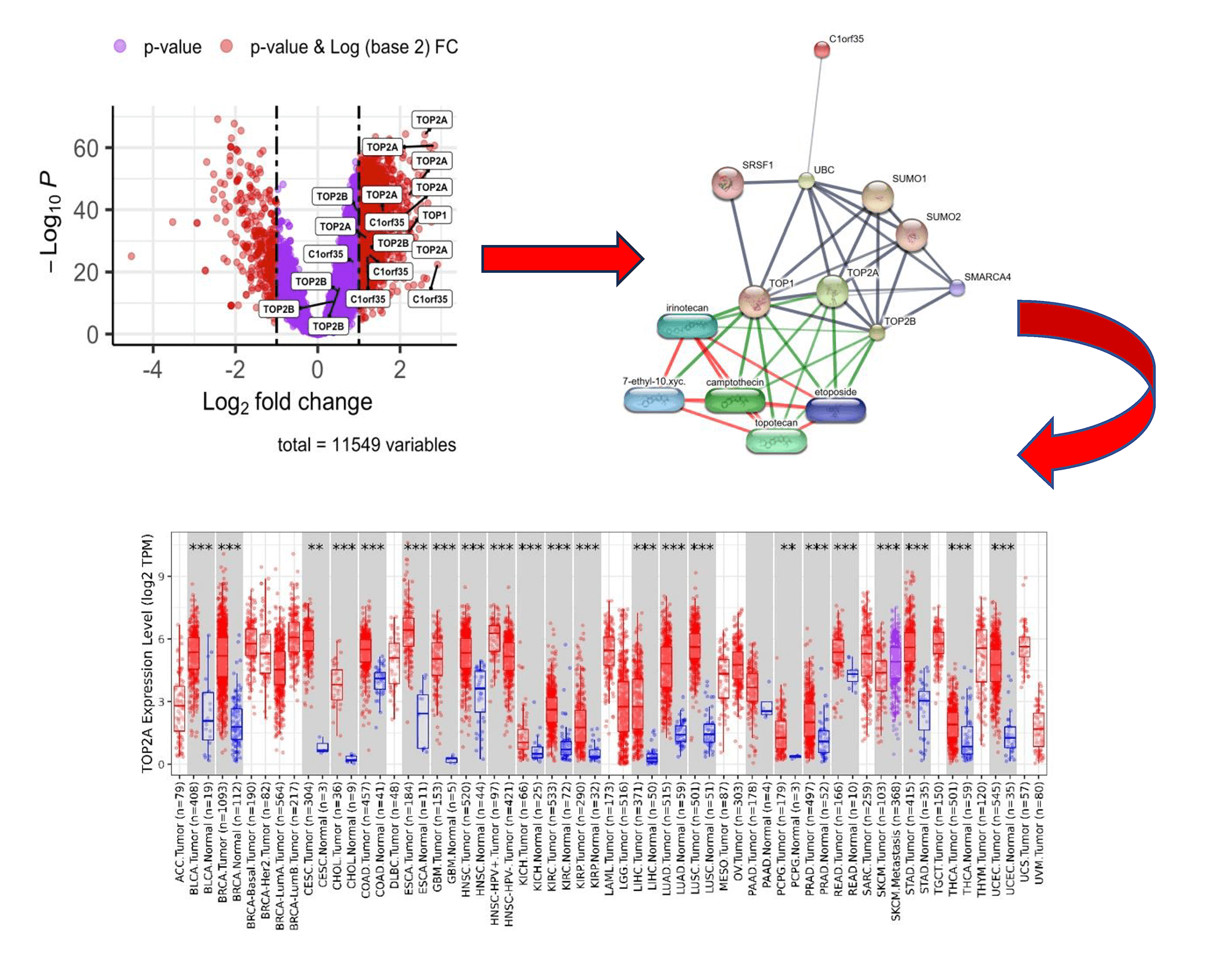
Objective: Hepatocellular carcinoma (HCC) is a leading cause of cancer-related mortality worldwide, largely due to late diagnosis, molecular heterogeneity, and limited prognostic biomarkers. Aberrant protein phosphorylation plays a critical role in cancer progression by regulating DNA damage response, cell cycle control, and signaling pathways; however, the prognostic relevance of phosphorylation events in key DNA topology–related proteins remains incompletely understood. This study aimed to investigate the prognostic significance of phosphorylation of TOP1, TOP2A, TOP2B, and C1orf35 in HCC and to characterize their associated molecular features to identify potential diagnostic and therapeutic biomarkers.
Methods: Publicly available HCC phosphoproteomic and proteomic datasets were analyzed to identify significantly upregulated phosphorylation sites of TOP1, TOP2A, TOP2B, and C1orf35. Integrated bioinformatics and machine learning approaches were applied, including Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), protein–protein interaction (PPI) network analysis, drug–gene interaction analysis, and survival analyses (overall and disease-free survival).
Results: A total of 11,547 phosphorylation sites corresponding to 4043 phosphoproteins were quantified from 159 HCC patients. Phosphorylation of TOP1, TOP2A, TOP2B, and C1orf35 was significantly upregulated. Enriched pathways included DNA damage response, homologous recombination repair, cell cycle regulation, SUMOylation, and TP53 signaling. PPI analysis identified these proteins as highly interconnected hub nodes. Elevated expression was significantly associated with poor clinical outcomes.
Conclusions: Phosphorylated TOP1, TOP2A, TOP2B, and C1orf35 are strongly associated with HCC progression and poor prognosis, highlighting their potential as prognostic biomarkers and therapeutic targets. These insights not only enhance our understanding of the complex molecular mechanisms underlying HCC but also offer promising avenues for the identification of novel therapeutic targets.
Graphical Abstract
Keywords
Hepatocellular carcinoma (HCC); phosphoproteomics; proteomics; bioinformatics; the Kaplan-Meier method; nano–liquid chromatography coupled with electrospray ionization tandem mass spectrometry (nLC-ESI-MS/MS); c-BioPortal
 Open Access
Open Access